Structural Research of Host-Defense Proteins
Host-defense proteins play a pivotal role in the innate immune response against invading pathogens, such as DNA viruses and bacteria. Among these essential proteins, STING (Stimulator of interferon genes) has emerged as a key adaptor protein involved in innate immunity. Extensive structural research has provided fascinating insights into the activation mechanism of STING and its potential applications in vaccines and cancer immunotherapies.
The groundbreaking work in the field of structural biology has allowed scientists to gain a deeper understanding of STING-mediated immunity. The initial challenge was to resolve the high-order oligomers formed by STING upon activation. The cGAMP-induced STING oligomers were challenging to analyze due to their weak resolution. However, a significant breakthrough came with the discovery of compound 53 (C53), a small molecular agonist that promotes STING oligomerization and activation through a mechanism distinct from cGAMP. The cryo-EM structure of STING bound to both C53 and cGAMP revealed a stable oligomer formed through side-by-side packing with a curled overall shape. The binding of C53 to a cryptic pocket in the STING TM domain induced outward shifts of TM helices, mediating the formation of the high-order oligomer. Together, these findings shed light on the intricate structural changes underlying STING activation.
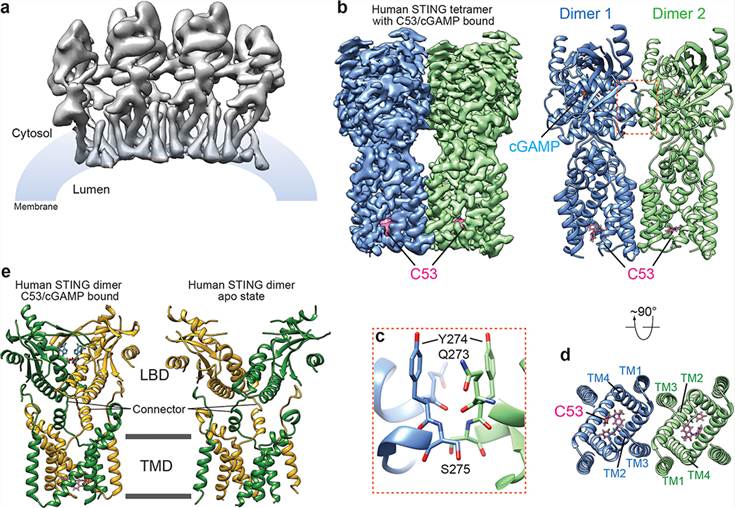 Figure 1. Structure of the high-order oligomer of human STING bound to both cGAMP and C53. (Lu D, et al., 2022)
Figure 1. Structure of the high-order oligomer of human STING bound to both cGAMP and C53. (Lu D, et al., 2022)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| STING (aka TMEM173, MITA, ERIS, or MPHYS), full-length apo state (expressed in HEK293 cells) | Homo sapiens | Cryo-EM single particle analysis | 4.10 Å | 6NT5 |
| STING (aka TMEM173, MITA, ERIS, or MPHYS) bound to both cGAMP and Compound 53 (expressed in HEK293 cells) | Homo sapiens | Cryo-EM single particle analysis | 3.45 Å | 7SII |
| STING (aka TMEM173, MITA, ERIS, or MPHYS) bound to HB3089 (expressed in HEK293 cells) | Homo sapiens | Cryo-EM single particle analysis | 3.47 Å | 8GT6 |
| adapter protein AP-1 in complex with STING (phosphorylated) (expressed in Escherichia coli) | Homo sapiens | Cryo-EM single particle analysis | 2.34 Å | 7R4H |
| STING (aka TMEM173, MITA, ERIS, or MPHYS) apo state (expressed in HEK293 cells) | Gallus gallus | Cryo-EM single particle analysis | 4.00 Å | 6NT6 |
| Complex between human TBK1 and chicken STING (expressed in HEK293 cells) | Homo sapiens, Gallus gallus | Cryo-EM single particle analysis | 3.30 Å | 6NT9 |
Table 1. Structural research of host-defense proteins.
As a renowned industry leader in structural biology, Creative Biostructure has been at the forefront of providing cutting-edge structural analysis services for host-defense proteins like STING. With years of experience, our team of highly skilled experts is dedicated to advancing the understanding of immune defense mechanisms through state-of-the-art technologies and methodologies.
Creative Biostructure offers a wide range of advanced techniques, including X-ray crystallography, nuclear magnetic resonance (NMR) spectroscopy, and cryo-electron microscopy (cryo-EM), to unravel the three-dimensional structures of host-defense proteins. Contact us to explore the potential of our capabilities, which can strengthen your research efforts and bring you one step closer to attaining your scientific objectives. Allow us to act as the driving force behind your progress and success.
References
- Lu D, et al. Activation of STING by targeting a pocket in the transmembrane domain. Nature. 2022, 604(7906): 557-562.
- Xie Z, et al. Structural insights into a shared mechanism of human STING activation by a potent agonist and an autoimmune disease-associated mutation. Cell Discovery. 2022, 8(1): 133.
- Liu Y, et al. Clathrin-associated AP-1 controls termination of STING signalling. Nature. 2022, 610(7933): 761-767.