Structural Research of Cyclooxygenases
Prostaglandin endoperoxide H synthases (PGHSs), also known as cyclooxygenases (COXs), are bifunctional enzymes with both cyclooxygenase and peroxidase activities and are key enzymes that catalyze the conversion of arachidonic acid to prostaglandins.
X-ray crystallographic analysis has proven to be a powerful tool for studying COX active sites and revealing COX inhibitor binding patterns.
The three-dimensional structure of sheep subtype prostaglandin H2 synthase 1 has been determined by X-ray crystallographic studies. The protein consists of three separate folding units, the epidermal growth factor structural domain, the membrane binding pattern and the enzyme structural domain. The active site of the cyclooxygenase is formed by a long hydrophobic channel, which is also the binding site for nonsteroidal drugs. Based on the strong conformation of the membrane-binding pattern, the enzyme is a single membrane protein that is integrated into only one lobe of the lipid bilayer.
COX-1 is a key target for a large number of drugs used clinically to treat a variety of human diseases. the sheep isoform of COX-1 (oCOX-1) has been studied a dozen times crystallographically in complex with inhibitors and substrates. More difficult is the use of the human isoform (hCOX-1). A large number of folded and post-translationally modified hCOX-1 can be expressed using a baculovirus expression system for structural analysis. hCOX-1 crystals were subjected to X-ray diffraction experiments.
hCOX-1 shares very high sequence identity (92%) with oCOX-1 and, like the sheep enzyme, crystallizes in the hexagonal space group in the presence of lithium chloride and sodium citrate. hCOX-1 conformation is specific to two regions. The region between residues 270-290 is part of the loop around the heme site and does not appear to be directly involved in the electron transfer mechanism between prostaglandin G2 (PGG2) and the prostate moiety that occurs at this site. the region 70-110 links EGF to the membrane-bound structural domain and, therefore, is close to the substrate pocket.
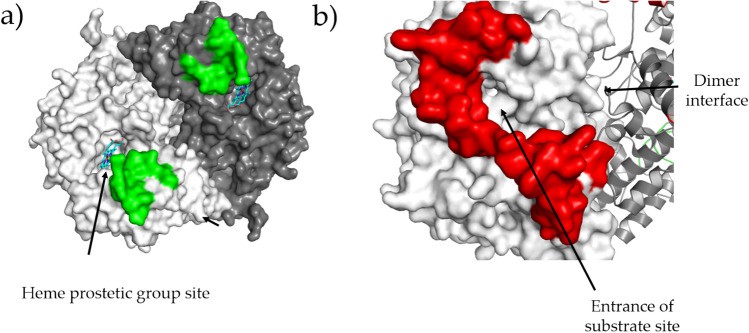 Figure 1. Regions whose conformation is peculiar for hCOX-1. (Miciaccia M, et al., 2021)
Figure 1. Regions whose conformation is peculiar for hCOX-1. (Miciaccia M, et al., 2021)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Ram Prostaglandin H2 synthase-1 | Ovis aries | X-ray diffraction | 3.50 Å | 1PRH |
| COX-1 in complex with bromoaspirin | Ovis aries | X-ray diffraction | 3.40 Å | 1PTH |
| COX-1 in complex with flurbiprofen | Ovis aries | X-ray diffraction | 3.10 Å | 1CQE |
| COX-1 in complex with ibuprofen | Ovis aries | X-ray diffraction | 2.61 Å | 1EQG |
| COX-1 in complex with flurbiprofen | Ovis aries | X-ray diffraction | 2.70 Å | 1EQH |
| COX-1 in complex with flurbiprofen methyl ester | Ovis aries | X-ray diffraction | 2.75 Å | 1HT5 |
| COX-1 in complex with alclofenac | Ovis aries | X-ray diffraction | 2.69 Å | 1HT8 |
| COX-1 in complex with O-actylsalicylhydroxamic acid | Ovis aries | X-ray diffraction | 3.20 Å | 1EBV |
| COX-1 in complex with alpha-methyl-4-biphenyl acetic acid | Ovis aries | X-ray diffraction | 2.00 Å | 1Q4G |
| COX-1 in complex with Flurbiprofen + Mn (III) PPIX cofactor. | Ovis aries | X-ray diffraction | 2.00 Å | 2AYL |
| R120Q/Native Cyclooxygenase-1 Heterodimer mutant in complex with flurbiprofen | Ovis aries | X-ray diffraction | 2.75 Å | 3N8W |
| COX-1 in complex with nimesulide | Ovis aries | X-ray diffraction | 2.75 Å | 3N8X |
| Aspirin Acetylated COX-1 in complex with Diclofenac | Ovis aries | X-ray diffraction | 2.60 Å | 3N8Y |
| COX-1 in Complex with flurbiprofen | Ovis aries | X-ray diffraction | 2.90 Å | 3N8Z |
| Ram Prostaglandin H2 synthase-1 (expressed in Spodoptera frugiperda) | Ovis aries | X-ray diffraction | 3.05 Å | 3N8V |
| COX-1 in complex with the subtype-selective derivative 2a (expressed in Spodoptera frugiperda) | Ovis aries | X-ray diffraction | 3.35 Å | 7JXT |
| Ram Prostaglandin H2 synthase-1 (expressed in Spodoptera frugiperda) | Homo sapiens | X-ray diffraction | 3.36 Å | 6Y3C |
| Cyclooxygenase-2 | Mus musculus | X-ray diffraction | 3.00 Å | 1CX2 |
| Cyclooxygenase-2 with bound indomethacin-ethylenediamine-dansyl conjugate | Mus musculus | X-ray diffraction | 2.22 Å | 6BL4 |
| Cyclooxygenase-2 with bound indomethacin-butyldiamine-dansyl conjugate | Mus musculus | X-ray diffraction | 2.22 Å | 6BL3 |
| α-DOX | Arabidopsis thaliana | X-ray diffraction | 1.51 Å | 4HHR |
| α-DOX (expressed in E. coli) | Arabidopsis thaliana | X-ray diffraction | 1.70 Å | 4HHA |
Table 1. Structural Research of Cyclooxygenases.
X-ray crystallography is a technique that is widely used in structural biology. It is based on the principle of irradiating an X-ray beam onto a molecular crystal and then obtaining a high-precision image of the protein structure by measuring the resulting diffraction pattern. It is particularly suitable for analyzing the three-dimensional structure of membrane proteins.
If you need an accurate and comprehensive analysis and understanding of protein structure and function, choose Creative Biostructure's X-ray crystallography service. We use state-of-the-art X-ray crystallography equipment to provide high-resolution images of protein crystal structures, ensuring that you get the most accurate results. If you are interested in learning more about our protein structural analysis services, please contact us for more information.
References
- Picot D, et al. The X-ray crystal structure of the membrane protein prostaglandin H2 synthase-1. Nature, 1994, 367(6460): 243–249.
- Loll P J, et al. The structural basis of aspirin activity inferred from the crystal structure of inactivated prostaglandin H2 synthase. Nature Structural Biology, 1995, 2(8): 637–643.
- Garavito, R M et al. The 3.1 Å X-ray crystal structure of the integral membrane enzyme prostaglandin H2 synthase-1. Advances in prostaglandin, thromboxane, and leukotriene research, 1995 (23): 99-103.
- Selinsky B S, et al. Structural analysis of NSAID binding by prostaglandin H2 synthase: time-dependent and time-independent inhibitors elicit identical enzyme conformations. Biochemistry, 2001, 40(17): 5172–5180.
- Loll P J, et al. o-acetylsalicylhydroxamic acid, a novel acetylating inhibitor of prostaglandin H2synthase: Structural and functional characterization of enzyme-inhibitor interactions. Molecular Pharmacology, 2001, 60(6): 1407–1413.
- Gupta K, et al. The 2.0Å resolution crystal structure of prostaglandin H2 synthase-1: Structural insights into an unusual peroxidase. Journal of Molecular Biology, 2004, 335(2): 503–518.
- Gupta K, et al. 2.0 Å structure of prostaglandin H2synthase-1 reconstituted with a manganese porphyrin cofactor. Acta Crystallographica Section D Biological Crystallography, 2006, 62(2): 151–156.
- Sidhu R S, et al. Comparison of cyclooxygenase-1 crystal structures: Cross-talk between monomers comprising cyclooxygenase-1 homodimers. Biochemistry, 2010, 49(33): 7069–7079.
- Miciaccia M, et al. Three-dimensional structure of human cyclooxygenase (hcox)-1. Scientific Reports, 2021, 11(1).
- Kurumbail R G, et al. Structural basis for selective inhibition of cyclooxygenase-2 by anti-inflammatory agents. Nature, 1996, 384(6610): 644–648.
- Xu S, et al. Fluorescent indomethacin-dansyl conjugates utilize the membrane-binding domain of cyclooxygenase-2 to block the opening to the active site. Journal of Biological Chemistry, 2019, 294(22): 8690–8698.
- Goulah C C, et al. The crystal structure of α-dioxygenase provides insight into diversity in the cyclooxygenase-peroxidase superfamily. Biochemistry, 2013, 52(8): 1364–1372.
- Friedrich L, et al. Learning from nature: From a marine natural product to synthetic cyclooxygenase‐1 inhibitors by automated de Novo Design. Advanced Science, 2021, 8(16): 2100832.
