Structural Research of Paramyxoviridae
Members of the Paramyxoviridae family are enveloped viruses with an unsegmented negative-stranded RNA genome that causes different types of infections in vertebrates. Related diseases include measles, mumps, and respiratory infections. Some viruses, such as the highly pathogenic zoonotic Nipah virus (NiV) and Hendra virus (HeV), cause fatal epidemics. Despite the availability of targeted vaccines, there is an urgent need for effective antiviral treatment for post-exposure prophylaxis.
Structural Information of Paramyxoviruses
Analysing the three-dimensional delicate structure of viruses as well as the structure and molecular assembly mechanism of ribonucleoprotein complexes in viruses is essential for an in-depth understanding of the biological properties of viruses, vaccine design, and antiviral drug discovery. With the rapid development of cryo-electron tomography (cryo-ET) and cryo-electron microscopy (cryo-EM), researchers have analyzed the structures of virus particles of several members of Paramyxoviridae. The structures show that the virus particles are enveloped, mostly spherical, and can produce filamentous virus particles. All members have two membrane glycoprotein complexes, the attachment protein (H, HN, or G) and the fusion protein (F), which appear as spikes on the surface of the viral particle. Matrix proteins within the envelope stabilize the viral structure. The nucleocapsid core consists of genomic RNA, nucleocapsid proteins, phosphoproteins, and polymerase proteins.
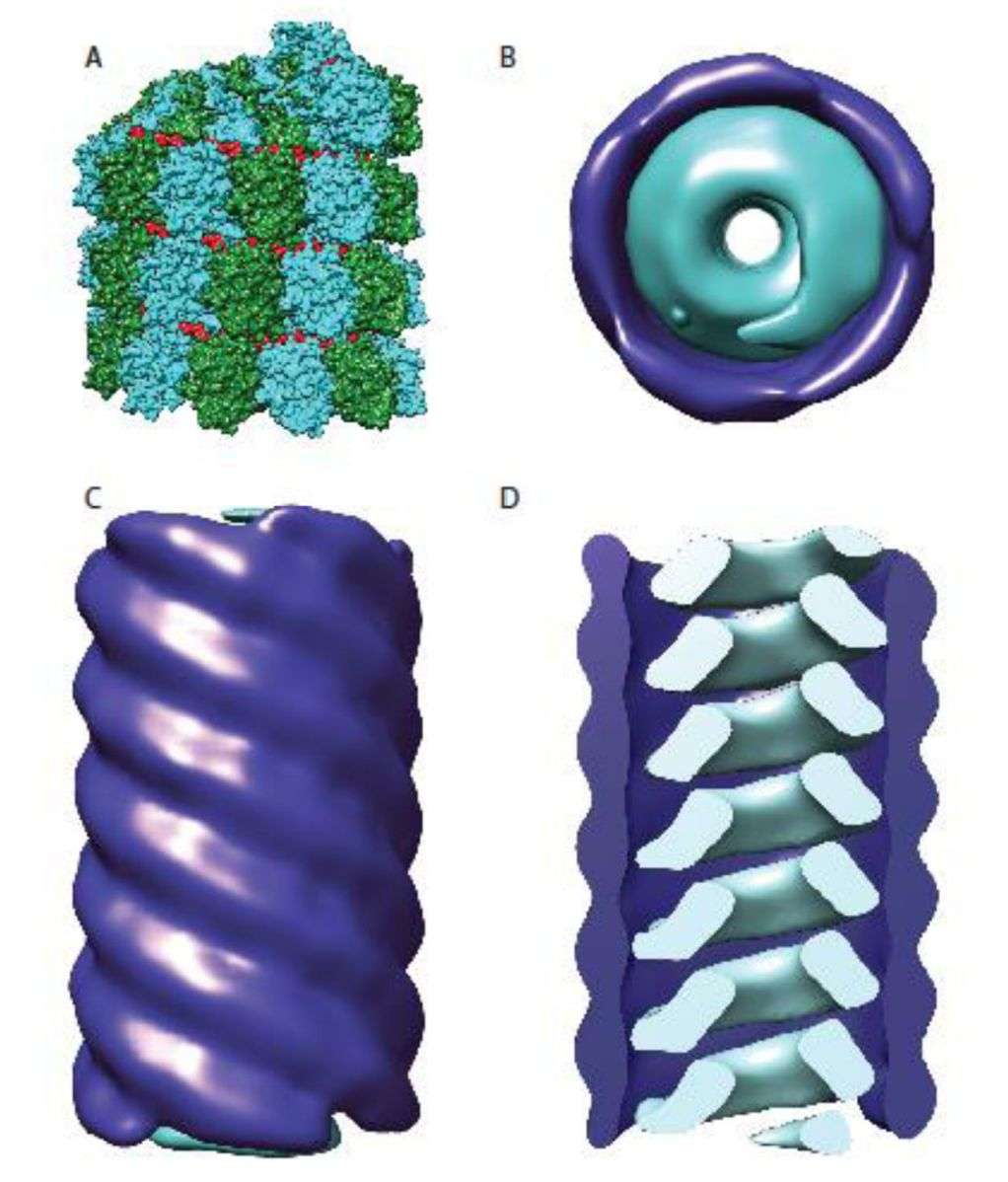 Figure 1. Organization of the paramyxovirus nucleocapsid. (Cox RM, Plemper RK, 2017)
Figure 1. Organization of the paramyxovirus nucleocapsid. (Cox RM, Plemper RK, 2017)
Advances in Structural Research on Paramyxovirus Proteins
The process of viral entry is a major focus of vaccine and drug development. With the discovery and structural characterization of new members, attention has been drawn to the major targets of neutralizing antibodies on the surface of paramyxoviruses. Researchers have studied NiV and HeV and found two surface glycoproteins (attachment proteins and fusion proteins), that facilitate the interaction between the virus and host cells. The attachment protein is a single type II transmembrane protein displayed as a tetramer, consisting of a dimer of the homodimer. The fusion protein is a type I viral fusion protein that assembles into a homotrimer.
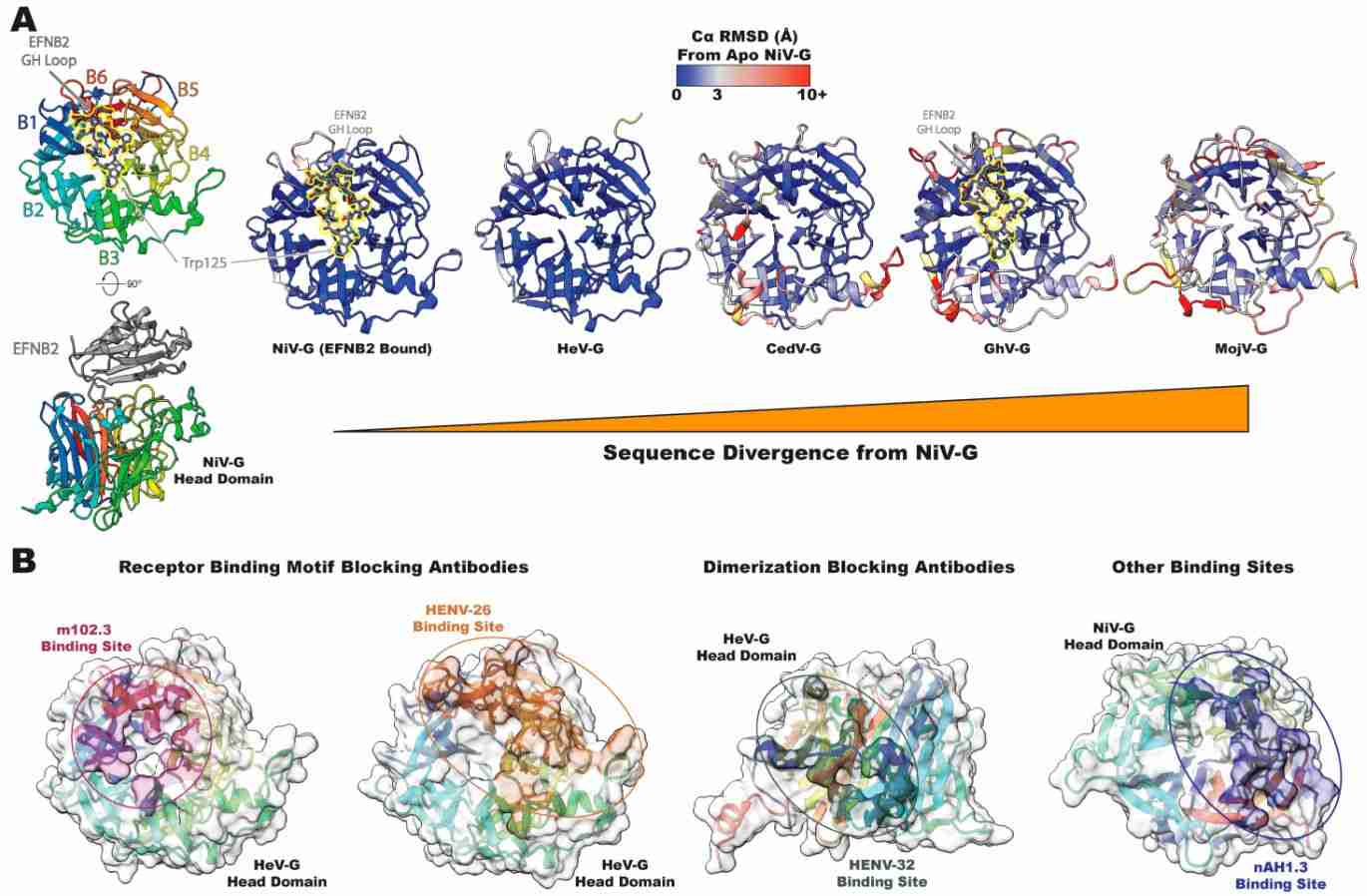 Figure 2. NiV and HeV head domain structure and antigenicity. (May AJ, Acharya P., 2024)
Figure 2. NiV and HeV head domain structure and antigenicity. (May AJ, Acharya P., 2024)
Structural biology is often the basis for vaccine and drug development, and focusing on knowledge of paramyxovirus structural biology, including areas such as antigenicity, conformational dynamics, and fusion-promoting mechanisms, is essential for the development of effective defense and control strategies.
Creative Biostructure, as a leader in structural analysis of biological macromolecules, offers a comprehensive range of virus-like particles (VLPs) products and solutions for viral structure analysis. Our VLPs are safe and non-infectious tools for vaccine research, providing researchers with an efficient platform for vaccine and antiviral drug discovery.
| Cat No. | Product Name | Virus Name | Source | Composition |
| CBS-V559 | Canine Distemper Virus VLP (M Proteins) | Canine distemper virus | Mammalian cell recombinant | M (Matrix Protein) |
| CBS-V613 | Hendra virus VLP (M; AP3B1-derived polypeptides Proteins) | Hendra virus | Mammalian cell recombinant | M; AP3B1-derived polypeptides |
| CBS-V631 | Human parainfluenza virus type 3 VLP (M Proteins) | Human parainfluenza virus | Mammalian cell recombinant | M |
| CBS-V693 | Nipah virus VLP (F; G; M Proteins) | Nipah virus | Mammalian cell recombinant | F; G; M |
| Explore All Paramyxoviridae Virus-like Particle Products | ||||
Creative Biostructure is dedicated to research in viral structural biology. Our scientists have a deep understanding of the structural resolution of viral particles and VLPs based on knowledge of virology and long-term project experience. We have cutting-edge cryo-electron microscopy (cryo-EM) to characterize nanoscale VLPs by accurately determining viral morphological features and confirming particle integrity.
In addition, we work closely with clients to provide customized structural analysis strategies for viruses of interest. For a detailed quote, please feel free to contact us.
References
- Cox RM, Plemper RK. Structure and organization of paramyxovirus particles. Curr Opin Virol. 2017. 24: 105-114.
- May AJ, Acharya P. Structural Studies of Henipavirus Glycoproteins. Viruses. 2024. 16(2): 195.
- Bloyet LM. The Nucleocapsid of Paramyxoviruses: Structure and Function of an Encapsidated Template. Viruses. 2021. 13(12): 2465.