Mempro™ Nanodisc Computational Studies
Creative Biostructure has been committed to the development of nanodisc technology for years and developed an innovative and comprehensive Mempro™ Nanodisc platform to promote the research of membrane proteins and their complexes. Complementary to experiments, we have developed various Mempro™ Nanodisc models powered by molecular dynamics simulations, and we have developed a multiscale Mempro™ Nanodisc computational strategy composed of AT-MD and CG-MD simulations in order to study the detailed insights into structure and dynamics of both the lipid and the membrane proteins. We believe that the computational studies, which is a promising approach in the future scientific research, will greatly facilitate the developments of nanodisc.
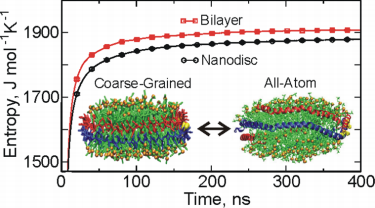 Figure 1. The configurational entropy of lipid in the nanodisc and a lamellar bilayer phase by using coarse-grained and all-atom simulations. (J. Phys. Chem. B, 2015)
Figure 1. The configurational entropy of lipid in the nanodisc and a lamellar bilayer phase by using coarse-grained and all-atom simulations. (J. Phys. Chem. B, 2015)
-
Mempro™ Nanodisc Model Development
As we know that there are many limited factors in experiments to study the dynamics of the assembly of nanodiscs, therefore, Creative Biostructure utilizes Mempro™ Nanodisc computational studies to overcome these problems. Obviously, the construction of various nanodisc models containing different phospholipids and membrane scaffold proteins (MSPs) is a prerequisite for further understanding of lipid-protein interactions and protein-protein interactions.
-
All-atom Mempro™ Nanodisc Dynamic Simulation
Creative Biostructure possesses a very mature technique of all-atom Mempro™ nanodisc dynamic simulation. We can perform AT-MD simulations which were carried out with a variety of GROMOS force fields. Compared with the experiments, AT-MD simulations can observe those missing details of thermodynamic, structural and dynamic properties of lipid and membrane proteins, and the resolution can be achieved at the atomic level. Along with these advantages, All-atom Mempro™ nanodisc dynamic simulation was adopted as an effective assay to study structural and dynamical properties of nanodiscs.
-
Coarse-grained Mempro™ Nanodisc Dynamic Simulation
Although all-atom simulation provides high resolution structural details of various nanodiscs, the time scale is limited to a few tens of nanoseconds or shorter somehow. To overcome this shortcoming, Creative Biostructure has established an advanced coarse-grained dynamic simulation method for the biological studies of membrane proteins and make sure the observed results are in an equilibrium state. In our CG-MD simulations, four non-hydrogen atoms are represented by one single interaction center, and the MARTINI force-field is used to describe CG-MD system. Compared with AT-MD simulation, CG-MD simulation can extend the simulation timescale quite a lot, which is more favorable for each nanodisc system to reach the equilibrium state. Furthermore, the simulation systems of nanodiscs can be much more larger in CG-MD.
Creative Biostructure also provides numerous Mempro™ Nanodisc products and services besides computational studies. Please feel free to contact us for a detailed quote.
References:
A. Debnath, et al. (2015). Structure and Dynamics of Phospholipid Nanodiscs from All-Atom and Coarse-Grained Simulations. Journal of Physical Chemistry B, 119(23): 6991-7002.
M. Vestergaard, et al. (2015). Bicelles and Other Membrane Mimics: Comparison of Structure, Properties, and Dynamics from MD Simulations.Journal of Physical Chemistry B, 119: 15831−15843.
M. A. Schuler, et al. (2013). Nanodiscs as a new tool to examine lipid-protein interactions. Methods in Molecular Biology, 974: 415-433.
