Structural Research of Sulfate-Uptake Transporters/Permeases
Sulfate transporters/permease (SulP) is an integral membrane protein that controls the passage of sulfate (SO42-) across the membrane lipid bilayer into the cellular and subcellular compartments. In all organisms, sulfate uptake is the first step in assimilation, a highly endoergic, ATP-requiring process. It is tightly controlled at the transcriptional level and is also regulated by post-translational modifications. Structural elucidation of the SulP family of proteins can further deepen the understanding of the action mechanism of SulP involved in transport.
Advances in SulP protein structure research
Research has shown that all SulPs are homodimers. Two of the subunits do not function independently. The low-resolution structure of the bacterial SulP transporter protein reveals dimer stoichiometry that is stabilized by its transmembrane core and mobile intracellular structural domains. The structure suggests that large movements of the STAS structural domain underlie the conformational changes that occur during transport. In addition, eukaryotic SulP proteins are typically larger than their prokaryotic homologs, and these proteins have 10-13 putative transmembrane α-helical spanners (TMS).
Structural analysis of the SulP protein CysZ
The research found that CysZ is described as a SO42- permease. Researchers gained insight into the molecular mechanism of CysZ-mediated SO42- transmembrane translocation based on crystal structure information, SO42- binding, and flux experiments. The crystal structure shows that CysZ is an α-helical integral membrane protein with two long transmembrane (TM) helices (H2b and H3a) and two pairs of shorter helices (H4b-H5a and H7-H8), which are only partially inserted into the membrane and form a funnel or tripod shape within the membrane. The protein has an extramembranous hydrophilic "head" containing two short helices, H1 and H6, and an iridescent arrangement of kinked helices H3b, H4a, and H5b. The helices H4b and H5a are partially inserted into the membrane, with the angle between them pointing towards the trifold axis in the center of the hexamer.
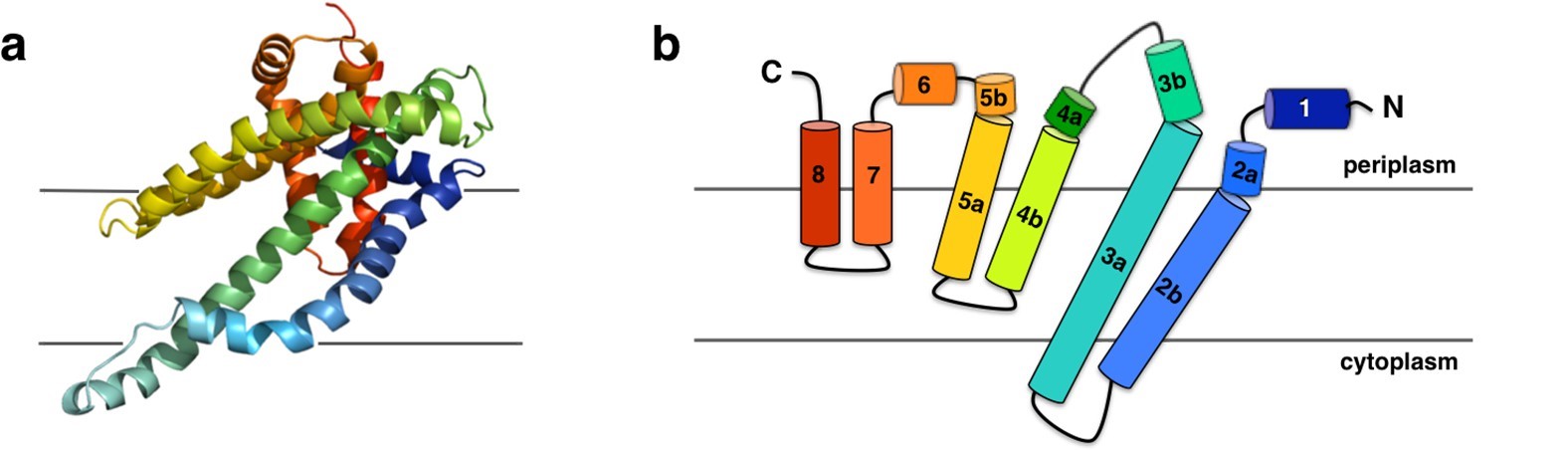 Figure 1. Structure and topology diagram of the P. denitrificans CysZ (PdCysZ) protomer. (Assur Sanghai Z, et al., 2018)
Figure 1. Structure and topology diagram of the P. denitrificans CysZ (PdCysZ) protomer. (Assur Sanghai Z, et al., 2018)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| CysZ | Idiomarina loihiensis | X-ray diffraction | 2.3 Å | 3TX3 |
| Sulfate transporter family protein | Wolinella succinogenes | X-ray diffraction | 1.85 Å | 3OIR |
| SulP transporter STAS domain | Wolinella succinogenes | X-ray diffraction | 1.6 Å | 4DGF |
| STAS domain of motor protein prestin | Rattus norvegicus | X-ray diffraction | 1.57 Å | 3LLO |
| STAS domain of RV1739c | Mycobacterium tuberculosis variant bovis | SOLUTION NMR | / | 2KLN |
| SLC26 transporter SLC26Dg in complex with a nanobody | Deinococcus geothermalis DSM 11300 | X-ray diffraction | 3.2 Å | 5DA0 |
| Transmembrane domain of the transporter SLC26Dg | Deinococcus geothermalis DSM 11300 | X-ray diffraction | 4.2 Å | 5IOF |
| Sulfate transporter AtSULTR4;1 | Arabidopsis thaliana | Cryo-EM single particle analysis | 2.75 Å | 7LHV |
| STAS domain of YchM bound to ACP | Escherichia coli | X-ray diffraction | 1.922 Å | 3NY7 |
| Prestin STAS domain | Gallus gallus | X-ray diffraction | 2.3 Å | 5EZB |
| 50S ribosome bound to antibiotic analog SLC26 | Escherichia coli | Cryo-EM single particle analysis | 2.32 Å | 8E47 |
| SLC26A9 | Homo sapiens | Cryo-EM single particle analysis | 2.6 Å | 7CH1 |
| STAS domain in complex with iodide | Rattus norvegicus | X-ray diffraction | 2.403 Å | 5EUZ |
| STAS domain in complex with chloride | Rattus norvegicus | X-ray diffraction | 1.87 Å | 5EUU |
Table 1. Structural research of the sulfate-uptake transporters/permeases.
Increasing interest in the structure and function of proteins and other biomolecules has led to the need for high-quality structural analysis services, and Creative Biostructure is a leading provider of structural analysis services for biomolecules such as sulfate-uptake transporters/permeases by X-ray crystallography, cryo-electron microscopy (cryo-EM), and NMR spectroscopy.
We offer a full range of services from sample preparation to data collection and processing to generate high-resolution structures of complex biological systems. If you are passionate about structural research of proteins and would like to learn more about the services we offer, please feel free to contact us. Our team is available 24/7 to satisfy your research requirements and provide the best solution for your project.
References
- Assur Sanghai Z, et al. Structure-based analysis of CysZ-mediated cellular uptake of sulfate. Elife. 2018. 7: e27829.
- Takahashi H. Sulfate transport systems in plants: functional diversity and molecular mechanisms underlying regulatory coordination. J Exp Bot. 2019. 70(16): 4075-4087.
- Piłsyk S, Paszewski A. Sulfate permeases phylogenetic diversity of sulfate transport. Acta Biochim Pol. 2009. 56(3): 375-384.