Mempro™ Membrane Protein Technology
Creative Biostructure has been committed to the study of membrane protein during the past decades, and we have established an innovative and advanced Mempro™ Membrane Protein Platform to promote the research of membrane proteins, providing a comprehensive services from gene to structure. Mempro™ Membrane Protein Platform has been focused on all kinds development of novel techniques to purify and produce high-quality membranes with high-yield, to incorporate those membrane proteins into different lipid systems such as liposomes, virus-like particles and nanodiscs in order to keep them in their native states, to determine the crystallization structure with various methods, to investigate their functions both in vivo and in vitro, and to offer molecular dynamics studies of membrane proteins as a complementary approach.
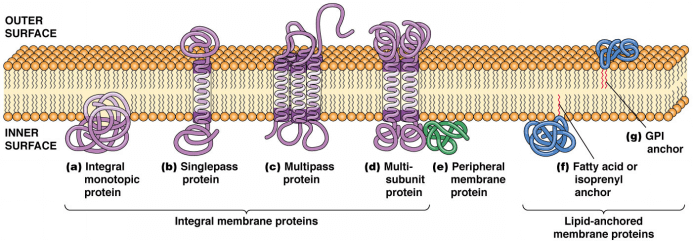 Figure 1. Various ways in which membrane proteins associate with the lipid bilayer.
Figure 1. Various ways in which membrane proteins associate with the lipid bilayer.
Membrane proteins are, as the name implies, located in the different membranes of the cell. In many cases they serve as gateways for molecules and signals into the cell making them of great relevance, for example as targets for pharmaceutical drugs. The membrane proteins are mainly classified into three types including integral membrane proteins, peripheral membrane proteins and lipid-bound membrane proteins. Integral membrane proteins are permanently embedded within the cell membrane. They are hardly removed from the lipid bilayer unless using detergents or nonpolar solvent which can destroy the cell membrane. Peripheral membrane proteins can be attached to the lipid bilayer or to integral membrane proteins temporarily by hydrophobic interaction, electrostatic interaction, and non-covalent interactions. They can be easily removed from the cell membrane. Lipid-bound membrane proteins are resided within the boundaries of the cell membrane entirely.
Membrane proteins can perform a wide range of functions which are vital to the cells:
Both inside-out and out-inside signal transductions between the internal and external of the cell,
Transportation of various molecules and ions through the membrane,
As activate enzymes,
As cell adhesion molecules for cell recognition and interactions involved in immune response.
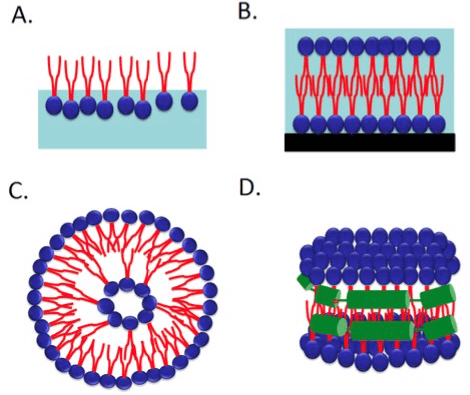 Figure 2. Schematic drawings of monolayer (A), lipid bilayer (B), liposome (C) and nanodisc (D). (Int. J. Mol. Sci., 2013)
Figure 2. Schematic drawings of monolayer (A), lipid bilayer (B), liposome (C) and nanodisc (D). (Int. J. Mol. Sci., 2013)
In recent years, much effort has made into elucidating the structure of transmembrane proteins. Many potential drug targets have been discovered among the different transmembrane protein families, such as the 7-TM receptors. Therefore, a huge interest in being able to perform drug screening on such proteins existed. Nevertheless, studying membrane proteins have proven to be a challenge due to the fact that they are especially difficult for purification and production. Creative Biostructure has rich experience in membrane protein purification and production with various expression systems such as insect cells system, yeast cells system, mammalian cells system, plant cells system and bacterial cells system. In order to reconstitute membrane proteins in model membrane systems in their native conformation after purification, Creative Biostructure has developed several novel platforms such as Mempro™ Virus-like Particles (VLPs) Platform, Mempro™ Liposome Platform, and Mempro™ Nanodisc Platform. These innovative model membrane systems are able to incorporate membrane proteins under mild conditions thus avoiding denaturing or modification of the protein, and able to be formed from different lipids to accommodate any membrane protein needs, and increase protein stability in solutions while preventing unwanted oligomerization and aggregation.
Based on the high-quality membrane proteins production, it is much easier for Creative Biostructure to developed an unparalleled UniCrys™ Crystallization and Structure Determination Platform to offer custom structural services for membrane proteins and their complex with native folding and functionality. Creative Biostructure provides a broad range of experimental methods in the study of tertiary membrane protein structure such as protein crystallography, nuclear magnetic resonance (NMR spectroscopy), small-angle s-ray and neutron scattering (SAXS and SANS), electron crystallography and single particle reconstruction, etc. Our rich-experienced scientists have successfully solved tons of crystallization structures of various membrane proteins.
Membrane proteins are extensively involved in protein-protein interactions, protein-lipid interactions, receptors, transporters, and ligand-binding, etc. Creative Biostructure can provide various quantitative and qualitative methods to analysis those membrane proteins, such as surface plasma resonance (SPR), isothermal titration calorimetry (ITA), and fluorescence resonance energy transfer (FRET), etc.
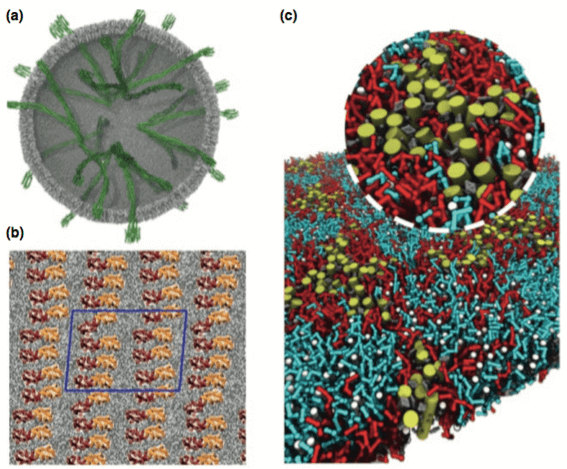 Membrane protein aggregation investigation by molecular dynamics studies. (Current Opinion in Structural Biology, 2013)
Membrane protein aggregation investigation by molecular dynamics studies. (Current Opinion in Structural Biology, 2013)
It plays as an invaluable tool for molecular simulation to better understand membrane proteins. With the improvements of both hardware and methods, we are allowed to access the physiologically relevant timescales and able to perform the simulation with large multimeric complexes in the system. As a complementary tool to study the structure and function of membrane proteins, Creative Biostructure has developed an advanced computational platform to provide molecular dynamics (MD) simulations composed of coarse-grained (CG) and all-atom (AA) simulations. With the increasing structure expansion of membrane proteins, we believe that the promising computational studies will greatly facilitate the way to elucidate the relationship between protein structure and function by saving the time and directing a better strategy for experimental studies
Creative Biostructure also provides a lot of other products and services based on the Mempro™ Membrane Protein Platform. Please see the related sections for detail information and feel free to contact us for a detailed quote.
References:
I. G. Denisov and S. G. Sligar. (2016). Nanodiscs for structural and functional studies of membrane proteins. Nature Structural & Molecular Biology, 23: 481–486.
L. Sercombe, et al. (2015). Advances and challenges of liposome assisted drug delivery. Frontiers in Pharmacology, 6: A286.
H. Shen, et al. (2013). Reconstitution of Membrane Proteins into Model Membranes: Seeking Better Ways to Retain Protein Activities. Int. J. Mol. Sci., 14: 1589-1607.
A. Zeltins et al., (2013). Construction and characterization of virus-like particles: a review. Mol Biotechnol., 53(1):92-107.