NMR Data Processing and Interpretation
NMR spectroscopy is one of the three approaches that enable determining 3D structures of bio-macromolecules, such as proteins, DNA/RNA, and their complexes, at atomic resolution. NMR could be used to study protein structure under its natural state. Another advantage is that it allows investigation of time-dependent chemical and conformational phenomena, including reaction and folding kinetics and intra-molecular dynamics, therefore, NMR plays a pivotal role within the life sciences.
NMR is based on the energy absorption and re-emission of the atom nuclei due to differences in an external magnetic field. Depending on the atom nuclei, different types of protein structural data are generated. NMR data processing from signal to 3D structure involves several steps (figure 1.) After collection of the primary experimental data, they are transformed to obtain spectra in which the individual frequency contributions or resonances. The resonances subsequently could be assigned to specific atoms. If enough resonances have been assigned, restraints can infer from the data. Pertaining to distances between atoms, dihedral domain orientations, etc. If an adequate number of restraints are available, these can be used to calculate the 3D structure. The derived structures represent the structure of the protein in solution.
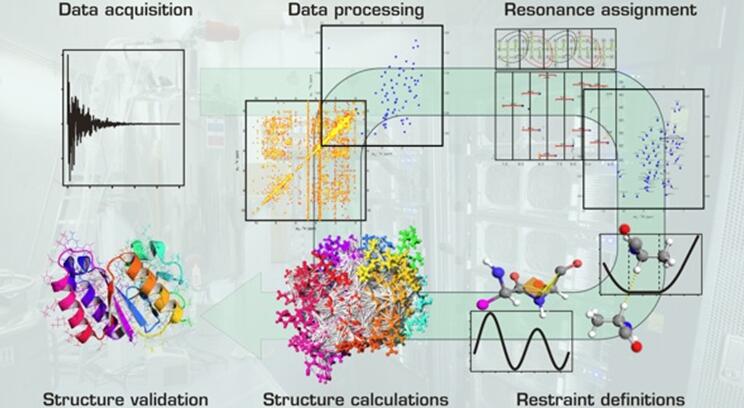 Figure 1. NMR data processing from signal to 3D structure
Figure 1. NMR data processing from signal to 3D structure
Software Used to Process and Interpret NMR Data:
Spectral processing
- Topspin2.1
- MNova
- NMRPipe/TALOS
- NUTS
Assignments and procheck
- Sparky
- CCPN
Molecular graphics
- PyMol
- MolMol
Databases and Resources:
- Biological Magnetic Resonance Bank (BMRB)
- Protein Data Bank (PDB)
- NMRShiftDB
Processing of NMR data has become a task for specialists, who are able to understand the data and their formats, as well as the software, programs, and database. Supported by state-of-the-art NMR platform and a team of specialists, Creative Biostructure dedicates to help customer with NMR experiments design, data collection and interpretation. Please feel free to consult with us through online inquiry.
Ordering Process
References
- Alonso A., Marsal S., and Julia A. (2015) Analytical methods in untargeted metabolomics: state of the art in 2015. Frontiers in Bioengineering and Biotechnology. 3: 23.
- Bahrami A., Assadi AH., Markley JL., and Eghbalnia HR. (2009) Probabilistic interaction network od evidence algorithm and its application to complete labeling of peak lists from protein NMR spectroscopy. PLoS Comput Biol 5(3): e1000307.

