Liposome Lamellarity Analysis
With years of experience in liposome research, scientists at Creative Biostructure are proud to provide comprehensive liposome products and services enabled by our advanced liposome platform. As a key parameter of liposomes, lamellarity is of great importance and is a nontrivial matter in the process of characterization. Our lamellarity analysis program intends to offer high quality services and products at highly competitive price to our worldwide clients.
Why lamellarity analysis is important to liposome?
Liposome lamellarity refers to the number of lipid bilayers of liposomes. Liposomes prepared by different methods may vary considerably in lamellarity. Lamellarity plays a crucial role in defining liposome properties: determining encapsulation efficiency, mediating diffusion rate of encapsulated agents out of liposomes, controlling drug release, penetration, etc. Moreover, lamellarity has significant effect on the intracellular fate of the drugs delivered by liposomes after they were taken up or processed in the cell.
Therefore, proper characterization of liposome lamellarity serves as a pivotal step in liposome preparation, and is the prerequisite for successful liposome-related applications.
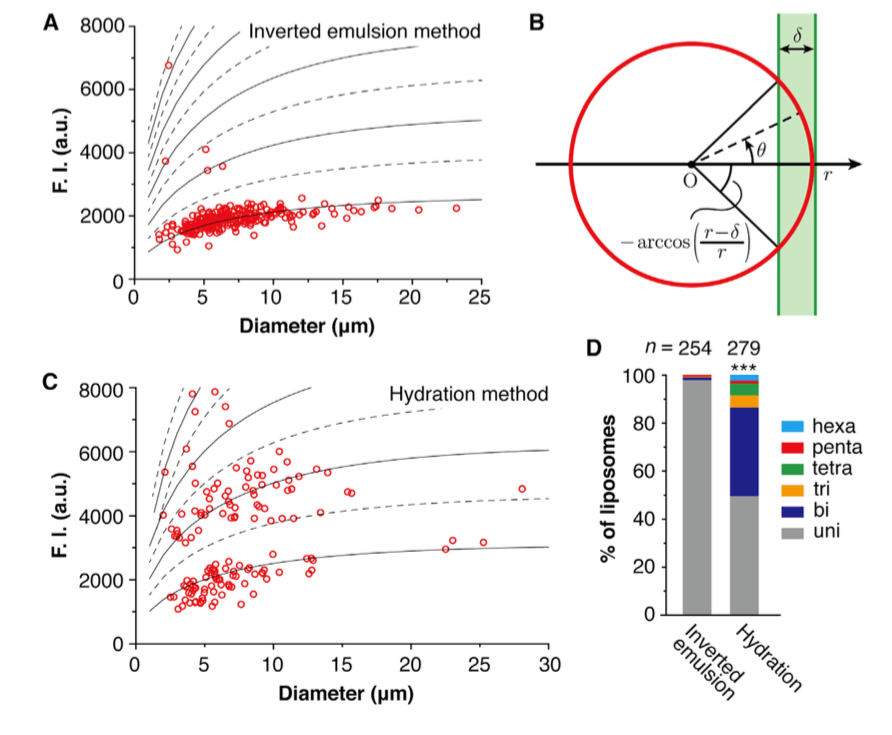 Figure 1. Lamellarity analysis of liposomes prepared by different methods. The liposomes with intensities that are below the lowest dashed line were characterized to be unilamellar, while the liposomes with intensities that are between the two lowest dashed lines were characterized to be bilamellar, and so on (A & C); A schematic diagram of a liposome under an epifluorescence microscope by side view (B); Lamellarity distributions of the liposomes prepared by different methods (D). (M. Chiba, et al., 2014)
Figure 1. Lamellarity analysis of liposomes prepared by different methods. The liposomes with intensities that are below the lowest dashed line were characterized to be unilamellar, while the liposomes with intensities that are between the two lowest dashed lines were characterized to be bilamellar, and so on (A & C); A schematic diagram of a liposome under an epifluorescence microscope by side view (B); Lamellarity distributions of the liposomes prepared by different methods (D). (M. Chiba, et al., 2014)
Liposome lamellarity determination methods
31P- nuclear magnetic resonance (NMR)
The determination of size and lamellarity of liposome is a direct application of nuclear magnetic resonance (NMR) in liposome quality control. In 31P-NMR, the external shift reagents including Mn2+, Co2+, and Pr3+ are widely used. The Mn2+ ion is a paramagnetic ion that interacts with the phospholipids located in the outermost of monolayer, leading to perturbations of the nuclear spin relaxation time. This is reflected on 31P-NMR spectrum as broadened peaks and reduced signal strength. The metal ion-induced signal loss can be quantified after data acquisition to calculate for lamellarity of the test sample. MLVs give rise to very broad peaks on 31P-NMR spectra due to the restricted anisotropic motion whereas SUVs tend to be less affected by such interaction.
Small angle X-ray scattering (SAXS)
Small angle X-ray scattering (SAXS) is another widely used technique for lamellarity determination. In a typical SAXS experiment, liposome is placed in glass capillary tubes, and then an X-ray radiation is applied to cause scattering curves of both the sample and the blank, which are recorded by a camera with a single-dimensional position sensitive detector. The Indirect Fourier Transformation provides the electron distance distribution p(r) which is calculated from scattered intensity in the measured sample. p(r) gives the radial contrast profile of Δ p(r) in electron density relative to the mean value, which could be used to solve the internal structure of the scattering particles. This method provides accurate information of liposome lamellarity.
Other methods
Other methods for quantitative determination of liposome lamellarity include label-free differential interference contrast (DIC) microscopy, epifluorescence microscopy, cryo-electron microscopy, and electron microscopy.
Creative Biostructure offers a full set of toolbox for liposome analysis and characterization, including all above mentioned methods, for liposome lamellarity measurement. Our service features in high cost effectiveness and rapid turnaround. A comprehensive documentation package will be generated for each project to deliver publication-ready images as well as experimental procedures. Please contact us to share more about your research and get a detailed quote.
Ordering Process
References
- M. Chiba, et al. (2014). Quantitative analysis of the lamellarity of giant liposomes prepared by the inverted emulsion method. Biophysical Journal, 107: 346-354.
- C. Chen, et al. (2013). Analytical techniques for single-liposome characterization. Analytical Methods, 5: 2150–2157.
- R. Suzuki, et al. (2014). Mono-cationic detergents play a critical role in the development of liposome-based gene vector via controlling its lamellarity. Nanoscale Research Letters, 16: 2227.
