Quick Comparison: Single Particle Cryo EM vs Tomography
In the rapidly evolving field of structural biology and nanotechnology, cryo-electron microscopy (cryo-EM) has emerged as a revolutionary technique for visualizing and understanding nanoscale structures. This powerful methodology encompasses several specialized approaches, with single particle analysis (SPA) and cryo-electron tomography (cryo-ET) being two primary methods that serve complementary purposes in structural analysis. While SPA excels at achieving atomic resolution through particle averaging, cryo-ET enables three-dimensional visualization of unique cellular structures in their native context. This comprehensive comparison explores their core differences, applications, and selection criteria to guide researchers in choosing the most appropriate technique for their specific needs.
Key Selection Factors at a Glance:
| Aspect | Single Particle Cryo-EM | Cryo-electron Tomography |
| Application Focus | Individual protein structures | In-situ molecular landscapes |
| Sample Type | Homogeneous, isolated particles | Heterogeneous, complex specimens |
| Resolution Range | Up to atomic resolution (~2-3Å) | 20-50Å (limited by cumulative electron dose) |
| Sample Preparation | Minimal manipulation | Requires precise sample thinning |
| Data Collection | Multiple particles, single images | Tilt series of one area |
| Image Processing | Particle averaging and 3D reconstruction | Tomographic reconstruction |
| Field of View | Limited to single particles | Larger field capturing cellular context |
| Key Strength | High-resolution molecular structures | Native cellular context |
Core Differences of Single Particle Cryo-EM and Tomography
Fundamental Principles of Each Technique
Cryo-EM represents a groundbreaking approach to structural biology, where specimens are preserved in their native state through rapid vitrification and examined under cryogenic conditions. This preservation method prevents the formation of ice crystals and maintains biological structures in their near-native state, offering unprecedented insights into molecular and cellular architecture.
Within the cryo-EM field, two major methodological approaches have emerged to address different analytical needs:
The Foundation of Single Particle Analysis (SPA)
SPA specializes in determining high-resolution structures of purified proteins and macromolecular complexes. This technique captures thousands of 2D images of identical particles frozen in random orientations. Through sophisticated computational processing, these images are aligned and averaged to generate detailed 3D reconstructions, often achieving atomic resolution (2-3Å). While SPA requires sample purification and homogeneity, it excels in revealing intricate molecular details crucial for understanding protein function and drug development.
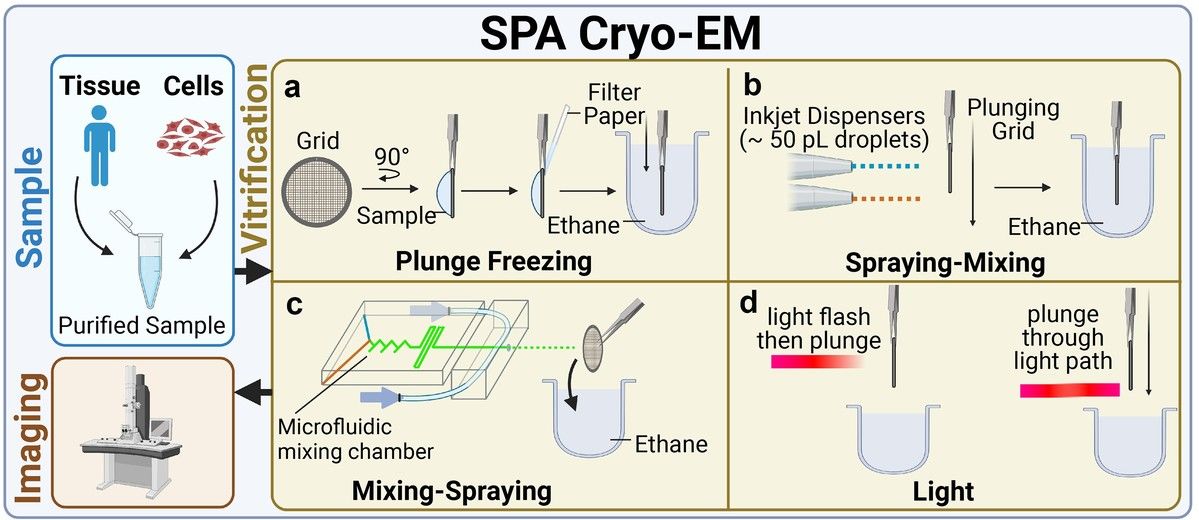 Figure 1. An overview of the typical workflow of single-particle analysis Cryo-EM. SPA cryo-EM workflow includes sample extraction, purification, and vitrification by plunge freezing or alternative methods like spraying-mixing, mixing-spraying, and time-resolved cryo-EM to capture short-lived states, followed by imaging. (Guaita M, et al., 2022)
Figure 1. An overview of the typical workflow of single-particle analysis Cryo-EM. SPA cryo-EM workflow includes sample extraction, purification, and vitrification by plunge freezing or alternative methods like spraying-mixing, mixing-spraying, and time-resolved cryo-EM to capture short-lived states, followed by imaging. (Guaita M, et al., 2022)
The Foundation of Cryo-Electron Tomography (Cryo-ET)
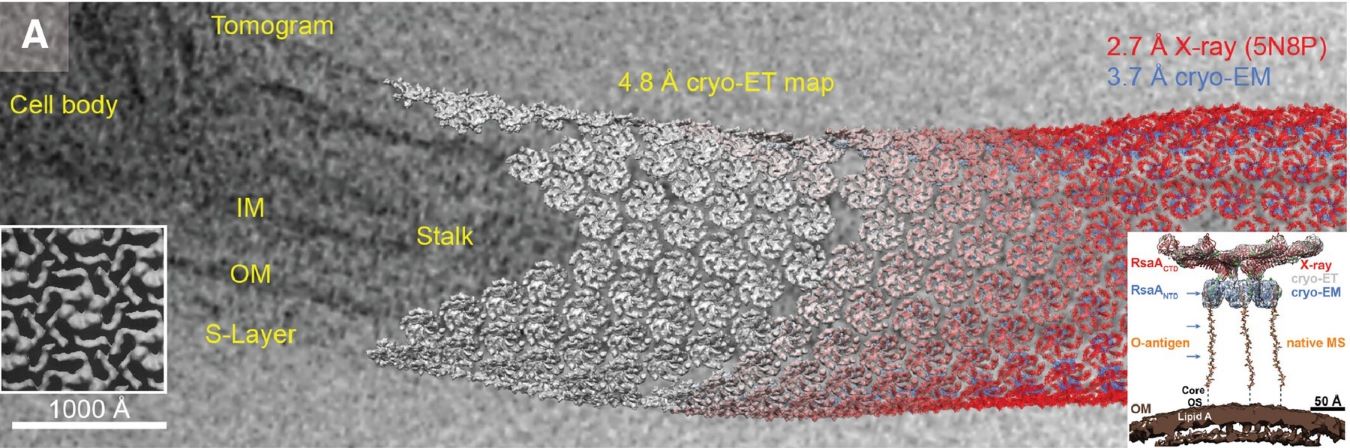
As a specialized application of cryo-EM, cryo-ET focuses on visualizing cellular structures in their native context. This method captures a series of 2D images at different tilt angles (typically from -60° to +60°) of a single specimen area, which are then computationally reconstructed into a 3D volume or 'tomogram'. While resolution is typically lower (20-50Å) due to cumulative electron dose constraints, cryo-ET uniquely reveals macromolecular organizations within the cellular environment. Sample preparation often requires sophisticated techniques like cryo-focused ion beam milling to achieve optimal specimen thickness.
Subtomogram Averaging
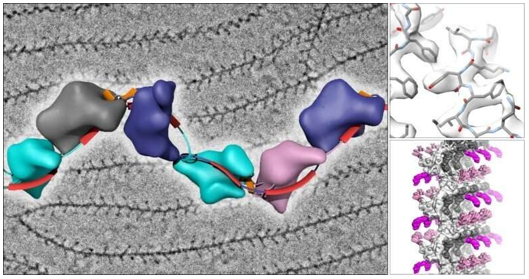
This advanced data processing technique bridges the gap between SPA and traditional tomography. By aligning and averaging multiple copies of similar structures from tomograms, researchers can achieve higher resolution reconstructions while maintaining contextual information. This approach is particularly powerful for studying membrane proteins and cellular complexes in their native environment.
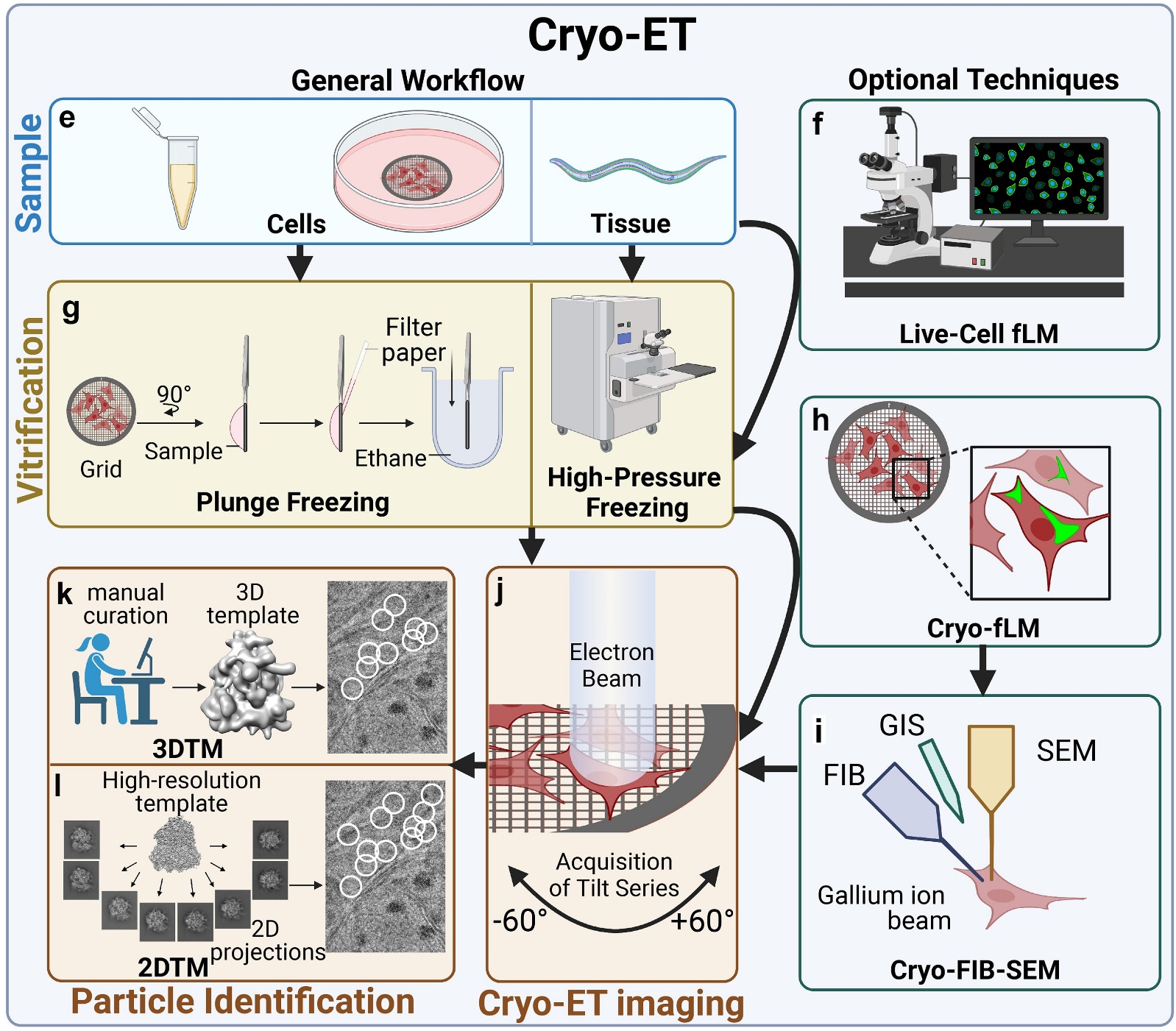 Figure 2. Cryo-ET workflow and optional techniques. Cryo-ET workflow involves vitrification by plunge freezing or high-pressure freezing, followed by imaging of thin specimens or targeted particles identified manually, with 3DTM or 2DTM. Optional techniques include live-cell fLM for optimal vitrification timing and cryo-fLM for locating tagged molecules, guiding cryo-FIB milling and imaging. (Guaita M, et al., 2022)
Figure 2. Cryo-ET workflow and optional techniques. Cryo-ET workflow involves vitrification by plunge freezing or high-pressure freezing, followed by imaging of thin specimens or targeted particles identified manually, with 3DTM or 2DTM. Optional techniques include live-cell fLM for optimal vitrification timing and cryo-fLM for locating tagged molecules, guiding cryo-FIB milling and imaging. (Guaita M, et al., 2022)
Key Applications and Use Cases
| Single Particle Cryo-EM Applications | Cryo-ET Applications |
|
|
Choose Single Particle Cryo-EM when:
- Sample consists of multiple identical copies
- Atomic or near-atomic resolution is required
- Target has inherent symmetry
- High throughput analysis is needed
- Structural dynamics studies are planned
Choose Cryo-ET when:
- Sample is unique or heterogeneous
- Spatial context is crucial
- Studying cellular architecture
- Analyzing irregular structures
- Investigating particle-cell interactions
- Membrane organization studies
Select Service
Technical Comparison of Single Particle Cryo-EM and Cryo-ET
Resolution and Visualization Capabilities in Cryo-EM Technologies
- Single particle analysis: SPA has revolutionized structural biology by routinely achieving atomic resolution (2-4Å), enabling visualization of protein side chains and bound ligands. This remarkable resolution is achieved through sophisticated averaging of thousands of identical particles, making it ideal for detailed structural studies of purified macromolecular complexes.
- Cryo-ET: While typically achieving lower resolution (20-50Å), it offers unique insights into cellular landscapes. The resolution limitation primarily stems from the cumulative electron dose distribution across multiple tilt angles and the missing wedge effect inherent in tomographic data collection. However, through subtomogram averaging, researchers can enhance resolution to 8-15Å for repeated structures within cellular contexts.
Sample Preparation Requirements for Cryo EM vs Cryo ET
- Single particle cryo-EM: It demands highly purified, homogeneous samples with optimal concentration (typically 0.1-1 mg/mL) and minimal background interference. The vitrification process must produce ice thickness under 100 nm for optimal imaging conditions. This stringent requirement ensures consistent particle visualization and reliable 3D reconstruction.
- Cryo-ET: It offers more flexibility in sample preparation but introduces its own complexities. Cellular samples require careful vitrification and often need additional preparation steps such as focused ion beam milling to achieve appropriate specimen thickness (<500 nm). The ability to work with heterogeneous samples in their native cellular context is a unique advantage, though it requires sophisticated sample thinning techniques and careful handling to preserve structural integrity.
| Parameter | Single Particle Analysis (SPA) | Cryo-Electron Tomography (Cryo-ET) |
| Sample Thickness | <100 nm ideal | <500 nm, requires FIB milling for thicker samples |
| Concentration | 0.1-1 mg/mL typical | Variable, depends on cellular context |
| Sample Purity | High homogeneity required | Accepts heterogeneous samples |
| Sample State | Purified protein complexes | Native cellular context |
| Key Requirements | - High sample homogeneity - Minimal contaminants - Stable protein complexes |
- Suitable specimen thickness - Cellular integrity - Structural preservation |
Data Acquisition and Processing Workflows
- Single particle analysis: SPA involves collecting thousands of images across multiple grid areas, with each particle ideally representing a random orientation. Modern detectors enable motion correction and precise CTF estimation, crucial for high-resolution reconstruction. The processing pipeline involves particle picking, multiple rounds of classification, and sophisticated 3D refinement algorithms.
- Cryo-ET: Tomographic data collection follows a systematic tilt series acquisition, typically spanning -60° to +60° with precise dose fractionation across the series. This approach generates comprehensive 3D information about a single specimen area but requires complex alignment procedures and careful dose management. The processing workflow includes tilt series alignment, tomogram reconstruction, and optional subtomogram averaging for repeated structures.
| Parameter | Single Particle Analysis (SPA) | Cryo-Electron Tomography (Cryo-ET) |
| Acceleration Voltage | 200-300 kV | 200-300 kV |
| Electron Dose | 40-60 e-/Ų total | 100-150 e-/Ų distributed across tilt series |
| Imaging Strategy | - Multiple fields of view - Random orientations - Motion correction |
- Tilt series (-60° to +60°) - Dose fractionation - Focus gradient correction |
| Processing Steps | - Particle picking - 2D classification - 3D classification - 3D refinement - Validation |
- Tilt series alignment - Tomogram reconstruction - Subtomogram averaging - CTF correction - Final reconstruction |
Time Investment and Resource Considerations
- Single particle cryo-EM: Single particle analysis typically requires 2-3 days for sample optimization, followed by 24-72 hours of automated data collection. The subsequent processing pipeline can extend from one to several weeks, depending on sample complexity and desired resolution.
- Cryo-ET: Cryo-electron tomography projects often demand longer preparation times, particularly when FIB milling is required (3-5 days). While individual tilt series collection takes 1-2 hours, the comprehensive analysis of cellular volumes and subsequent subtomogram averaging can extend processing time to several weeks.
Both methods require substantial computational resources and expertise, with investments in high-performance computing infrastructure being essential for efficient data processing.
Application-Specific Decision Guide for Cryo-EM Methods
Decision Matrix for Method Selection
| Research Goal | Primary Method | Key Requirements | Alternative Approaches |
| Atomic Resolution Protein Structure | SPA | - Purified sample - Stable protein - Sufficient concentration |
X-ray crystallography NMR for small proteins |
| Cellular Architecture | Cryo-ET | - Thin samples - Intact cells - FIB-milling capability |
Correlative light microscopy Serial section EM |
| Drug-Target Interactions | SPA | - Stable complex - High homogeneity - Concentration optimization |
X-ray crystallography Cryo-ET for membrane proteins |
| Large Complex Assemblies | Combined Approach | - Method integration - Multiple expertise - Advanced computing |
Sub-tomogram averaging Hybrid methods |
For Protein Structure Studies:
- Resolution < 4Å needed → SPA
- Membrane protein → SPA, consider Cryo-ET
- Dynamic states → Combined approach
For Complex Assemblies:
- Homogeneous samples → SPA
- Heterogeneous states → Cryo-ET
- Context-dependent function → Combined approach
For Cellular Studies:
- Native context crucial → Cryo-ET
- Specific complex localization → Correlative microscopy
- Multiple conformations → Sub-tomogram averaging
For Drug Development:
- Binding site details → SPA
- Membrane targets → Cryo-ET
- Delivery mechanisms → Combined approach
Atomic-Resolution Protein Structure Determination
The quest for atomic-resolution protein structures predominantly favors single particle analysis when working with stable, homogeneous protein complexes larger than 50 kDa. This method excels in revealing molecular details crucial for understanding protein function, particularly for soluble proteins and symmetrical assemblies. Success depends heavily on sample quality optimization, including careful protein engineering for stability enhancement and buffer condition refinement. For membrane proteins or highly dynamic systems, researchers should consider implementing gradient fixation techniques or exploring complementary methods such as cryo-ET for context-dependent structural information.
Cellular Architecture and Organization Studies
Cryo-ET emerges as the definitive choice for investigating cellular architecture and protein complexes within their native environment. This method provides unique insights into cellular organization, membrane-associated structures, and macromolecular assemblies in situ. The approach particularly shines in analyzing cell-cell interactions and membrane protein organizations in their physiological context. Success hinges on meticulous sample preparation, including precise lamella preparation through focused ion beam milling, and often benefits from correlation with fluorescence microscopy for targeted structural analysis.
Drug Development and Therapeutic Design
In pharmaceutical research, the choice of cryo-EM methodology depends critically on the specific stage and requirements of drug development. Single particle analysis proves invaluable for high-resolution ligand binding studies and structure-based drug design, offering atomic details of drug-target interactions and conformational changes. For membrane protein drug targets or studies of drug delivery mechanisms, cryo-electron tomography provides crucial insights into cellular context and delivery system behavior. The integration of both methods often yields comprehensive understanding, from atomic-resolution binding site details to cellular distribution patterns.
Complex Macromolecular Assemblies
Large macromolecular complexes often demand a sophisticated hybrid approach, combining the strengths of both single particle analysis and cryo-electron tomography. This integrated strategy proves particularly valuable for assemblies exceeding 1 MDa or those with hierarchical organization. Single particle analysis can reveal high-resolution details of stable core structures, while cryo-ET provides essential contextual information about assembly organization and interactions. Success in these studies often requires careful consideration of complex stability, heterogeneity, and the specific resolution requirements for different components.
Advanced Method Integration Strategies
Modern structural biology increasingly benefits from the strategic integration of multiple cryo-EM approaches. Subtomogram averaging bridges the gap between single particle analysis and traditional tomography, particularly valuable for membrane proteins and cellular complexes. This integration requires sophisticated data collection strategies and computational approaches but offers unprecedented insights into structure-function relationships. Researchers should consider factors such as sample characteristics, required resolution, and available resources when designing integrated approaches.
Select Service
Related Reading
In conclusion, both cryo-ET and single particle cryo-EM (SPA) are powerful techniques that have revolutionized structural biology, each with unique strengths suited for different research objectives. Whether you need atomic-level resolution for isolated macromolecules or 3D insights into cellular architecture, these methods offer unparalleled opportunities to advance your studies.
At Creative Biostructure, we specialize in providing cutting-edge cryo-EM services, including SPA and cryo-ET. Our team of experts is dedicated to supporting your research with customized solutions and state-of-the-art technology. Contact us to discuss your project requirements and discover how we can assist in achieving your research goals!
References
- Doerr A. Cryo-electron tomography. Nature Methods. 2017, 14(1): 34-34.
- Stewart P L. Cryo-electron microscopy and cryo-electron tomography of nanoparticles. Wiley Interdisciplinary Reviews: Nanomedicine and Nanobiotechnology. 2017, 9(2): e1417.
- Turk M, Baumeister W. The promise and the challenges of cryo-electron tomography. FEBS letters. 2020, 594(20): 3243-3261.
- Guaita M, Watters S C, Loerch S. Recent advances and current trends in cryo-electron microscopy. Current opinion in structural biology. 2022, 77: 102484.

-1.jpg)