RdRp (SARS-CoV-2) Homogeneous Assay
SARS-CoV-2 is a plus-stranded RNA virus. Its replication is mediated by the multi-subunit replication/transcription complex of the viral nonstructural protein (NSP). The core component of this complex is the catalytic subunit (NSP12) of RNA-dependent RNA polymerase (RDRP). NSP12 itself has little activity, and its function requires cofactors including NSP7 and NSP8, which can increase the template-binding and continuous synthesis of RDRP.
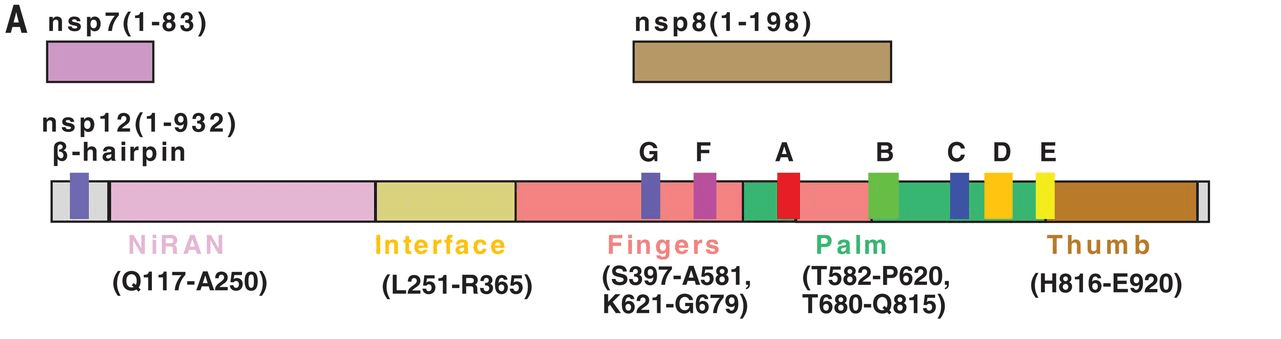 Figure 1. The schematic diagram for the components of the RdRp complex, containing nsp12, nsp7, and nsp8. The polymerase motif (A to G) and the β hairpin specific to SARS-CoV-2 are highlighted. (Wanchao Yin, et al. 2020)
Figure 1. The schematic diagram for the components of the RdRp complex, containing nsp12, nsp7, and nsp8. The polymerase motif (A to G) and the β hairpin specific to SARS-CoV-2 are highlighted. (Wanchao Yin, et al. 2020)
Creative Biostructure developed the SARS-CoV-2 RdRp Assay Kit based on the structure and function of these three nonstructural proteins. RdRp operates as a complex of NSP12, NSP7, and NSP8 proteins. The RdRp (SARS-CoV-2) Homogeneous Assay Kit is designed to measure the SARS-CoV-2 RNA-dependent RNA Polymerase (RdRp) activity for screening and profiling applications. Our RdRp Assay Kit is based on the RNA polymerization activity of the NSP12-NSP7-NSP8 complex in the presence of adenosine triphosphate (ATP) on polyuracil templates. So, the direct incorporation of ATP in the double-stranded RNA chain can be measured.
The key to the RdRp Homogeneous Assay Kit is a highly specific antibody that recognizes digoxigenin-labeled RNA Duplex. With this kit, only three simple steps on a microtiter plate are required for RdRp detection. First, a sample containing RdRp is incubated with the biotinylated substrate for one hour. Next, acceptor beads and primary antibodies are added, then donor beads, followed by reading the Alpha-counts.
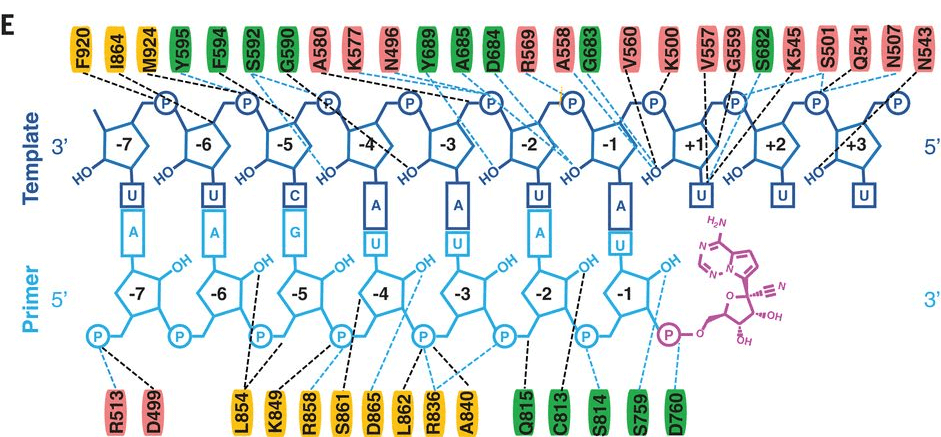 Figure 2. Diagram of detailed RNA interactions with RdRp. Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
Figure 2. Diagram of detailed RNA interactions with RdRp. Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
CONTRAINDICATIONS: Like other Homogeneous Assay Kits,green and blue dyes that absorb light in the AlphaScreen signal emission range (520-620 nm), such as Trypan Blue. Avoid the use of potent singlet oxygen quenchers such as sodium azide (NaN3) or metal ions (Fe2+, Fe3+, Cu2+, Zn2+, and Ni2+). The presence of >1% RPMI 1640 culture medium leads to a signal reduction due to the presence of excess biotin and iron in this medium. MEM, which lacks these components, does not affect AlphaScreen assays.
If you are interested in our RdRp (SARS-CoV-2) homogeneous assay service, please feel free to contact us. Creative Biostructure looks forward to cooperating with you.
Contact us to discuss your project!
Reference
- Wanchao Yin, et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020, doi:10.1126/science.abc1560.

 Figure 1. The schematic diagram for the components of the RdRp complex, containing nsp12, nsp7, and nsp8. The polymerase motif (A to G) and the β hairpin specific to SARS-CoV-2 are highlighted. (Wanchao Yin, et al. 2020)
Figure 1. The schematic diagram for the components of the RdRp complex, containing nsp12, nsp7, and nsp8. The polymerase motif (A to G) and the β hairpin specific to SARS-CoV-2 are highlighted. (Wanchao Yin, et al. 2020) Figure 2. Diagram of detailed RNA interactions with RdRp. Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
Figure 2. Diagram of detailed RNA interactions with RdRp. Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.