Protein-protein Interaction Network Analysis for Coronavirus Research
The ongoing outbreak of novel coronavirus (also termed as SARS-CoV-2) poses a challenge for public health. There is an urgent need to understand the molecular mechanism of this viral infection and to develop effective vaccines and antiviral drugs on this basis. Creative Biostructure can provide computational methods to support mapping protein-protein interaction (PPI) networks between coronaviruses and the host cells. This approach and the data generated from it can give a wealth of information about how the virus hijacks the host cell and cause disease and is of great significance for the development of viral vaccines and screening of antiviral drugs.
PPI Network Analysis of SARS-CoV-2
At the molecular level, viruses invade cells and manipulate them to replicate, survive, and cause disease. Given that they rely on human cells to complete their life cycle, viruses exploit cellular function is through PPIs within host cells. In turn, cells respond to viral infection by initiating immune responses that control and limit viral replication, which also depends on PPIs.
By studying the structural genomics and interactomics of novel coronavirus, the researchers reveal the functionally conserved regions of the evolutionary level of viral proteins. In this study, a comprehensive homology modeling analysis of 3D structures of SARS-CoV-2 proteins was carried out. The protein interaction complexes were predicted using model comparison, and a virus-host macromolecular interaction network was constructed, providing a protein structure reference for anti-coronavirus drug design. In addition, based on many biological experimental results, the researchers successfully mapped the PPI network between SARS-COV-2 and the human cell and analyzed the biological characteristics of the protein interaction map. According to the network, potential drugs that intervene in the protein interaction network have been screened, some of which enter clinical trials.
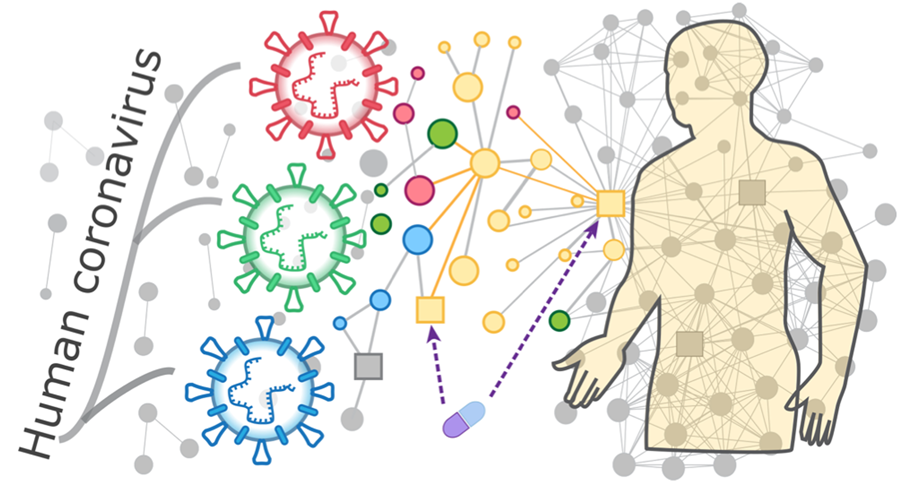 Figure 1. Schematic of PPI network analysis of human coronavirus. (Adapted from Zhou Y.; et al. 2020)
Figure 1. Schematic of PPI network analysis of human coronavirus. (Adapted from Zhou Y.; et al. 2020)
PPI Network Analysis Services for Coronavirus Research
Creative Biostructure specializes in PPI research, including computer-based and experimental protein interaction analysis services. In terms of PPI network analysis involved in coronavirus research supported by computing methods, we collect experimentally verified virus-host PPI data in public databases and integrate algorithms to establish the virus-host protein domain-domain interactions (DDI) database. And information of miRNA-target interaction can be integrated simultaneously. Based on the above data, a virus-host interaction network is constructed. By integrating the expression profile data of host tissues, the tissue-specific mechanism of viral infection, the difference of molecular mechanism of virus infection among different species, and the potential pathogenic mechanism of the virus are further investigated. The information about the general rules of virus-host interactions helps you for screening new antiviral drugs and development of vaccines to prevent coronavirus infections.
The study of the virus-host PPI network provides important clues for the selection of antiviral drug targets. We support the integration of viral protein structure data and small-molecule compound structure databases to achieve high-throughput virtual screening of virus-inhibiting compounds.
If you are interested in our coronavirus-related PPI network analysis services, please do not hesitate to contact us. Our customer service representatives are available 24 hours a day from Monday to Sunday.
Contact us to discuss your project!
References
- Cui H.; et al. Structural genomics and interactomics of 2019 Wuhan novel coronavirus, 2019-nCoV, indicate evolutionary conserved functional regions of viral proteins. BioRxiv. 2020.
- Gordon D E.; et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020: 1-13.
- Zhou Y.; et al. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discovery. 2020, 6(1): 1-18.

 Figure 1. Schematic of PPI network analysis of human coronavirus. (Adapted from Zhou Y.; et al. 2020)
Figure 1. Schematic of PPI network analysis of human coronavirus. (Adapted from Zhou Y.; et al. 2020)