Protein Sequence Analysis for Coronavirus Research
The human coronavirus pandemic poses a huge threat to global public health and the economy; however, no vaccine or specific antiviral drug has yet been approved to treat Coronavirus Disease 2019 (COVID-19) pneumonia. Therefore, the development of prevention and treatment approaches is especially important for the control of this emerging infectious disease. The analysis of viral proteins and host cell proteins can lay the foundation for understanding the mechanism of viral infection and achieving drug development. Creative Biostructure utilizes a variety of tools and techniques for analyzing and comparing protein sequences to help you increase understanding of characteristics, functions, domains, or evolution of proteins associated with coronavirus infection.
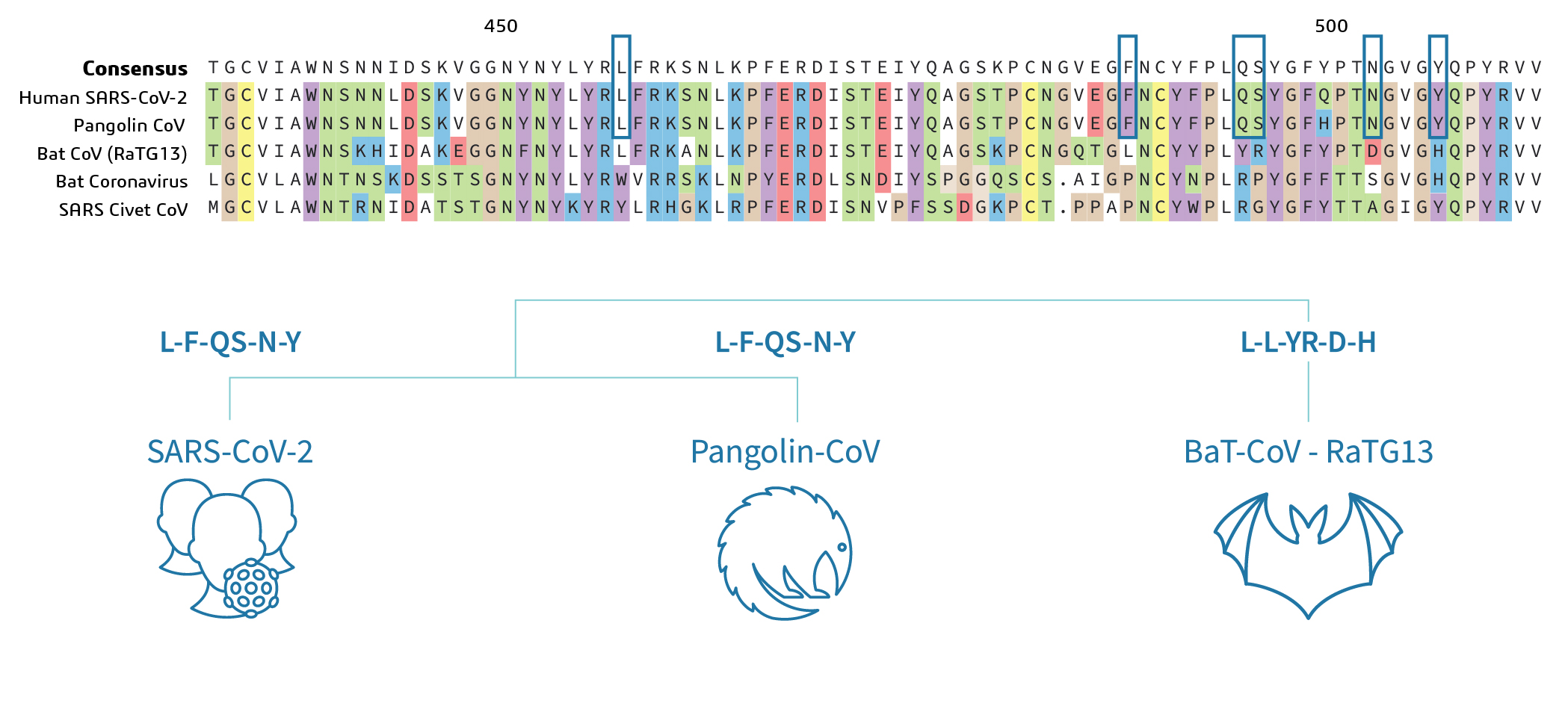 Figure 1. Multiple sequence alignment of Spike protein of Human, Pangolin and Bat SARS-CoV-2. (By Niranjani Iyer)
Figure 1. Multiple sequence alignment of Spike protein of Human, Pangolin and Bat SARS-CoV-2. (By Niranjani Iyer)
Protein Sequence Analysis Services for Coronavirus Research
Our professional bioinformatics analysis team provides the following protein sequence analysis services for coronavirus research, including but not limited to:
- Amino acid composition analysis: We can use single sequence analysis, multi-sequence analysis, and other techniques to provide amino acid composition analysis service.
- Protein motif prediction: Motif is an amino acid sequence pattern that symbolizes the signature of the protein family and has important biological significance. We can predict the protein motif for analyzing protein function.
- Signal peptide prediction: Signal peptides (SPs) present at the N-terminus of most newly synthesized proteins and contribute to the protein transport and localization. SPs can be predicted using a variety of methods, such as Neural network and Hidden Markov models (HMM).
- Transmembrane prediction: For transmembrane proteins, the transmembrane analysis is required, including the prediction of extracellular and intracellular regions and the number of the chain of membrane protein crosses the membrane.
- Binding sites predication of ligand: We can provide template-based, geometry-based, energy-based, propensity-based, and combination-based methods to predict protein-ligand binding sites according to the site-distinguishing properties.
Application of Protein Sequence Analysis in Coronavirus Research
- Compare protein sequences to find similarity and infer whether multiple proteins are homologous
- Identify differences and mutations between multiple protein sequences
- Identify molecular structure from sequence alone
- Identify intrinsic characteristics of the sequence, such as PTM sites, active sites, and regulatory elements
- Reveal protein evolution and diversity
Creative Biostructure’s protein sequence analysis service can provide extensive support for primary sequence analysis of viral and host cell proteins to facilitate coronavirus-related experimental research. Our customer service representatives are available 24 hours a day from Monday to Sunday, and you can contact us at any time to discuss your coronavirus-related project.
Contact us to discuss your project!

 Figure 1. Multiple sequence alignment of Spike protein of Human, Pangolin and Bat SARS-CoV-2. (By Niranjani Iyer)
Figure 1. Multiple sequence alignment of Spike protein of Human, Pangolin and Bat SARS-CoV-2. (By Niranjani Iyer)