Protein Homology Modeling for Coronavirus Research
The analysis of three-dimensional (3D) structure is of great importance for the mechanism of virus infection and the design of antiviral drugs. Although many breakthroughs have been made in structural biology techniques and structure determination methods such as X-ray crystallography, NMR spectroscopy and cryo-electron microscopy (cryo-EM) have been utilized to determine the structure of many biological macromolecules, the process of obtaining the 3D structure of the target protein is still difficult and time-consuming. Facing the continuous spread of the epidemic, we need to accelerate the understanding of the pathogenic mechanism of the virus. Creative Biostructure can generate structural models of coronavirus/host proteins through calculation methods, which will be a powerful alternative and supplement to the experimental method.
Application of Homology Modeling in Coronavirus Research
When the protein with unknown structure is similar to the protein with a known structure in the primary sequence, the latter can be used as a template to predict the 3D structure of the former through computer calculation and simulation. It has been shown that the protein 3D structure is evolutionarily more conserved than would be expected based on sequence conservation alone. Therefore, even if there are obvious differences in sequence but still have detectable similarities, the proteins will share common structural characteristics, especially the overall folding.
The novel coronavirus (SARS-CoV-2, formerly called as 2019-nCoV), which started outbreak in late December 2019, is homologous to SARS-CoV and MERS-CoV and belongs to the beta coronavirus. Sequence alignment of the SARS-CoV-2 genome sequence shows the gene sequences of SARS-CoV-2 and SARS-CoV are the most similar, with a homology of up to 79.5%. Considering the high sequence similarity, there have been some studies on homology modeling of certain key proteins of SARS-CoV-2 for rational drug design, such as main protease (Mpro), the Nsp12 RNA-dependent RNA polymerase (RdRp), Nsp13 helicase and chymotrypsin-like protease (3CLpro). Although research results based on computational methods lack experimental verification, structural information and identified potential hits can serve as a starting point for structure-based drug discovery.
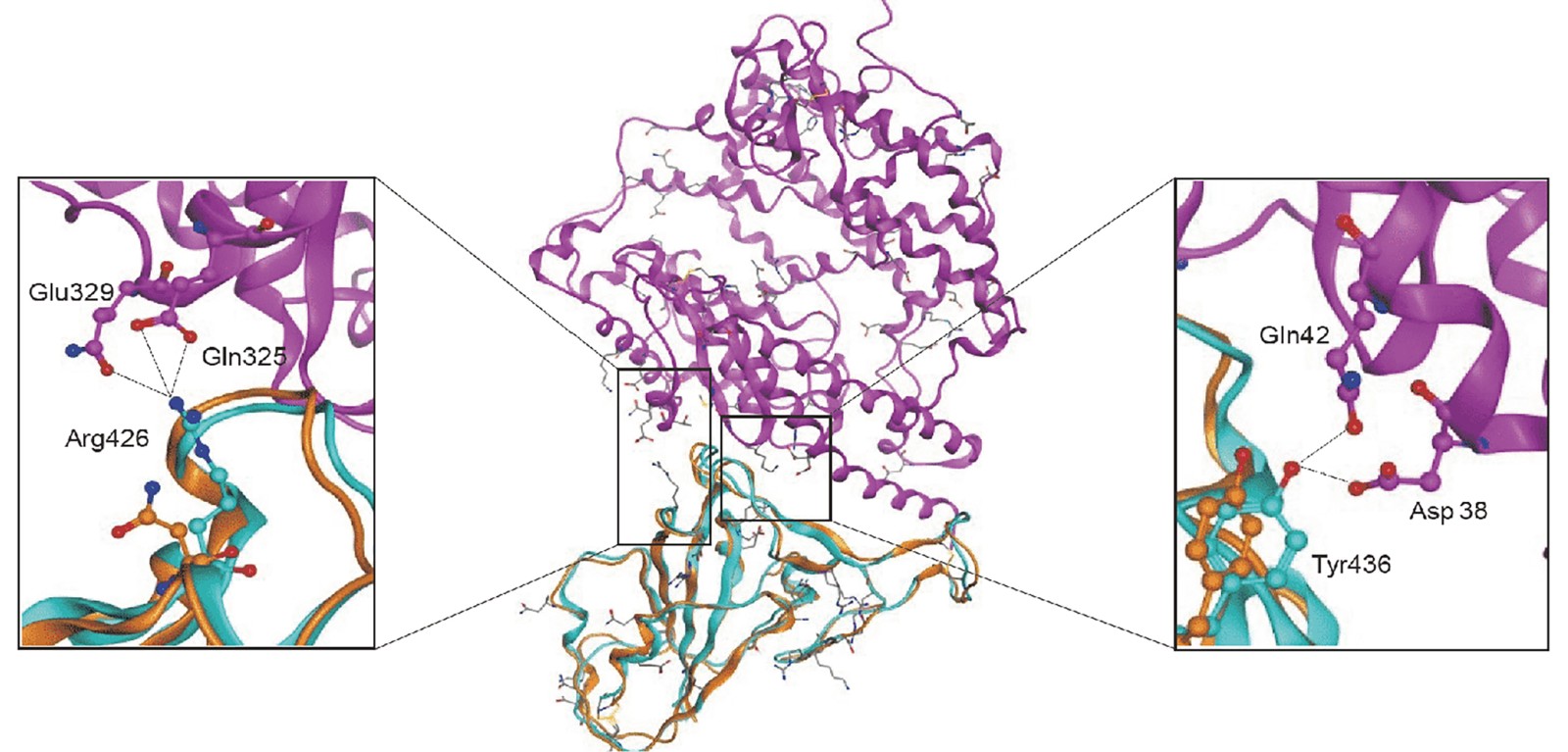 Figure 1. Structural modeling of the novel coronavirus S-protein complexed with human ACE2 molecule. (Adapted from Xu X.; et al. 2020)
Figure 1. Structural modeling of the novel coronavirus S-protein complexed with human ACE2 molecule. (Adapted from Xu X.; et al. 2020)
Protein Homology Modeling Services for Coronavirus Research
Creative Biostructure uses homology modeling to help you predict the 3D structure of related target proteins in coronavirus research. We have extensive experience in generating structure models of various types of proteins and protein complexes. The structure with low resolution generated by homology modeling contains enough information about the spatial arrangement of important residues in the protein and can guide the design of experiments, such as site-directed mutagenesis. The structural models we generated in the past are all quality validated and can be used for virtual screening and guidance of structure-based drug design during the antiviral drug discovery process.
The protein homology modeling services for coronavirus research provided by Creative Biostructure include integration of several recommended methods ranging from homology, ab-initio or threading. We can customize and perform the modeling according to the customer needs, and the client only needs to provide basic sequence information, such as the name of target protein, FASTA sequence, or sequence ID. Combination of modeling and short-term simulations under controlled parameters will ensure the reliability of the structure model for further studies.
Our customer service representatives are available 24 hours a day from Monday to Sunday, and you can contact us at any time to discuss your coronavirus-related project.
Contact us to discuss your project!
References
- Dong S.; et al. A guideline for homology modeling of the proteins from newly discovered betacoronavirus, 2019 novel coronavirus (2019‐nCoV). Journal of Medical Virology. 2020.
- Mirza M U, Froeyen M. Structural elucidation of SARS-CoV-2 vital proteins: Computational methods reveal potential drug candidates against main protease, Nsp12 polymerase and Nsp13 helicase. Journal of Pharmaceutical Analysis. 2020.
- Xu X.; et al. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Science China Life Sciences. 2020, 63(3): 457-460.

 Figure 1. Structural modeling of the novel coronavirus S-protein complexed with human ACE2 molecule. (Adapted from Xu X.; et al. 2020)
Figure 1. Structural modeling of the novel coronavirus S-protein complexed with human ACE2 molecule. (Adapted from Xu X.; et al. 2020)