The genomic DNA of higher organisms is compactly wrapped in the cell nucleus. DNA is tightly wound around a large number of histones called nucleosomes. For example, human cells contain about two meters of DNA in this way. However, genes must constantly undergo messenger RNA (mRNA) to guide protein synthesis. In addition, the entire DNA must be replicated before cell division, and DNA damage needs to be repaired. Therefore, cells must have methods to actively grant access to its genome.
This is exactly when the chromatin remodeler (also translated as a chromatin remodeling agent) works. Chromatin remodeling proteins play a vital role: they open DNA fragments by sliding back and forth on the nucleosomes, replace single histones, let DNA fragments release for transcription, and finally compress it when the transcription is complete. Given that all this occurs in a highly dynamic manner, chromatin remodeling proteins allow cells to respond quickly to changes in their environment – both for brewer’s yeast and human cells. When mediating gene accessibility, chromatin remodeling proteins are crucial for development and cell differentiation; cell types are determined by a set of genes that they express, and chromatin remodeling proteins help determine cell identity.
However, so far, little is known about what chromatin remodeling proteins look like and how they work. From a molecular perspective, functional chromatin remodeling proteins are often very large complexes containing many different protein components that coordinate their actions so that chromatin remodeling proteins resemble molecular machines. These features also make it very difficult to determine the detailed structure of chromatin remodeling proteins.
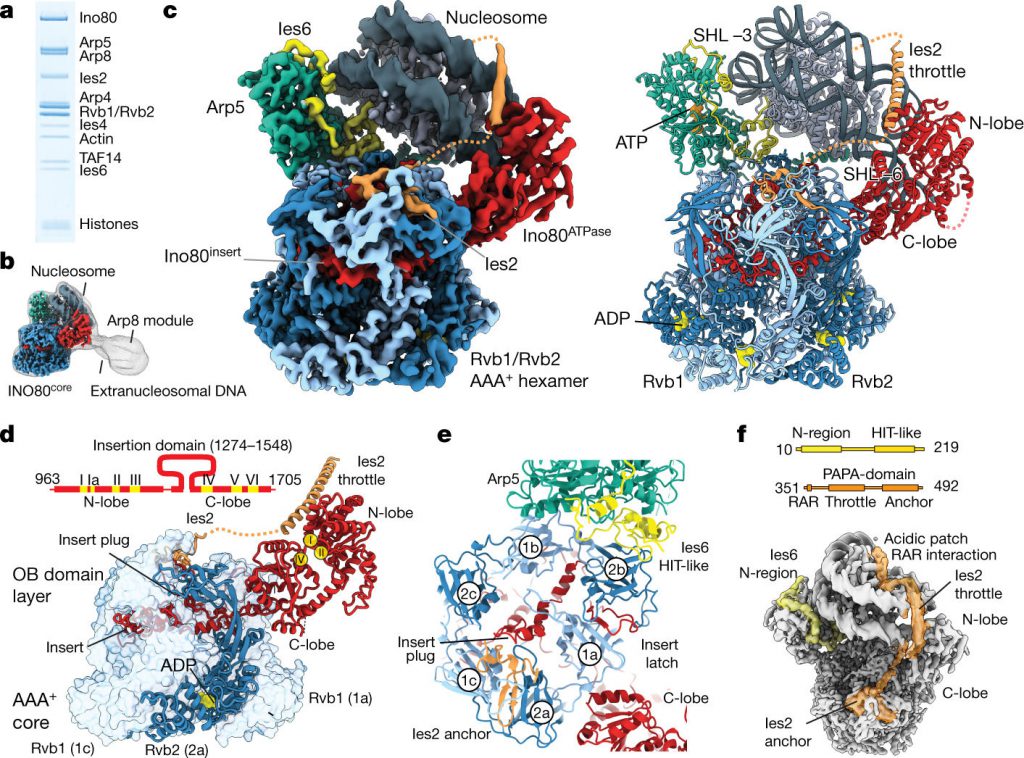
Now, in a new study, Professor Karl-Peter Hopfner, the director of structural biology at the Gene Center of the University of Munich, Germany, and his colleagues used cryogenic electron microscopy to reconstruct the three-dimensional structure of the chromatin remodeler INO80 (composed of 15 subunits) coming from Chaetomium thermophilum sliding on nucleosomes bonded to a single nucleosome. The results of the relevant study were published in Nature, April 19, 2018, and the paper is entitled “Structural basis for ATP-dependent chromatin remodelling by the INO80 complex”.
By analyzing the stochastically located view of the complex formed between the INO80 and the nucleosome in these cryo-EM images, Hopfner and his team stitched out the three-dimensional structure of the complex. This allowed them to reveal the complex interactions between this chromatin remodeling protein and DNA entangled on histones and to analyze how this entire molecular machine works.
In order to change the relative position of nucleosomes, INO80 must first weaken the contact between histones and DNA in the nucleosome. As a molecular motor subunit of INO80, the double-stranded DNA was detached from the nucleosome.
This molecular motor subunit sends DNA into the nucleosome. This results in the transient formation of double-stranded DNA loops, which may be an important intermediate for complex chromatin remodeling reactions on nucleosomes. On the one hand, this double-stranded DNA exposes several histones that can be replaced by other histones to form a different type of nucleosome. On the other hand, this double-stranded DNA eventually passes through another subunit, after which INO80 acts like a ratchet, allowing the nucleosome to “move” on the DNA. In this dismantling process, other subunits in INO80 support and stabilize part of the exposed nucleosomes.
In another study published in the same issue of the Nature, Dale B. Wigley, Xiaodong Zhang, and colleagues at the Imperial College in England presented the three-dimensional structure of the chromosomal remodeling agent INO80 when combined with a nucleosome. This structure reveals that human INO80 interacts with nucleosomes in a manner not previously described. The results of the relevant study were published in Nature, April 19, 2018, and the paper is entitled “Structure and regulation of the human INO80-nucleosome complex”.
Reference
Sebastian Eustermann, Kevin Schall, Dirk Kostrewa et al. Structural basis for ATP-dependent chromatin remodelling by the INO80 complex. Nature, 19 April 2018, 556:386–390, doi:10.1038/s41586-018-0029-y
Rafael Ayala, Oliver Willhoft, Ricardo J. Aramayo et al. Structure and regulation of the human INO80–nucleosome complex. Nature, 19 April 2018, 556:391–395, doi:10.1038/s41586-018-0021-6