To develop drugs that target specific cell surface proteins, it is helpful to understand other proteins in its vicinity. The pathology of many diseases can be understood by elucidating local biomolecular networks or microenvironments. To this end, the enzymatic proximity labeling platform is widely used to map a wider range of spatial relationships in subcellular structures. However, there has long been a search for techniques that can map the microenvironment more precisely.
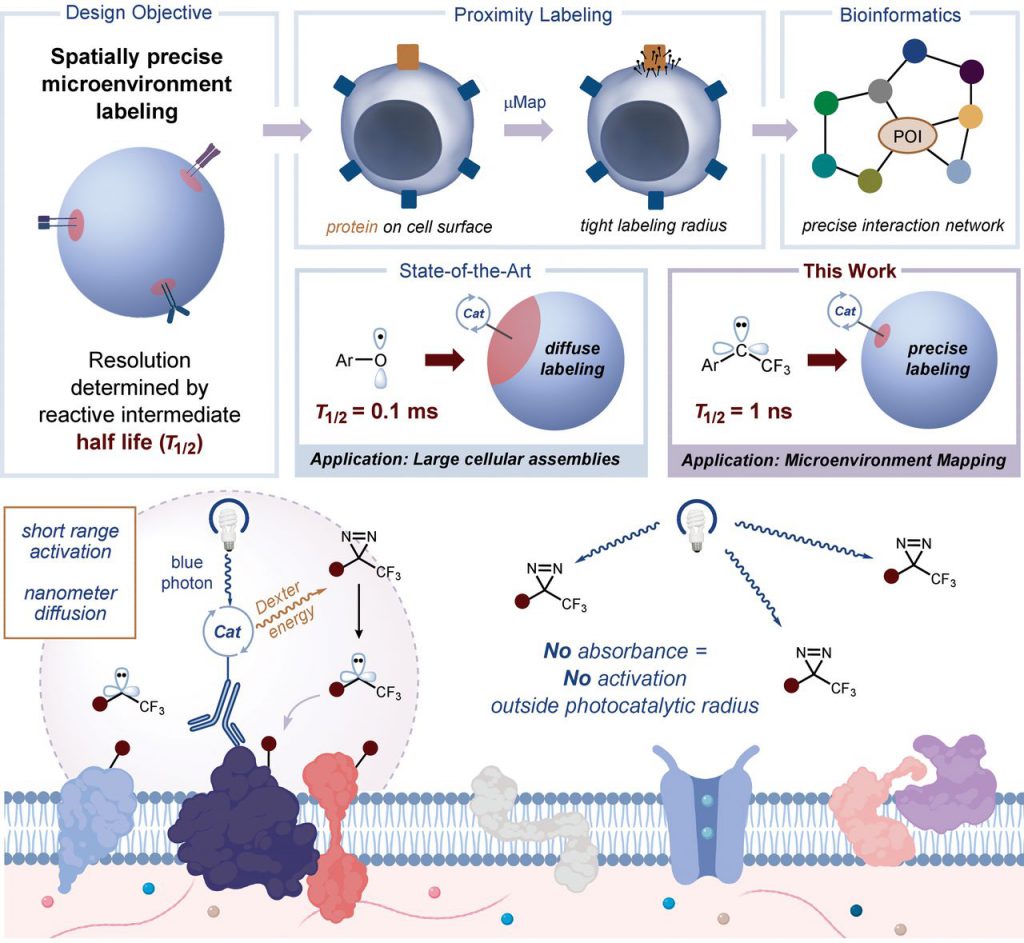
Geri et al describe a microenvironmental profiling platform that uses photocatalytic carbon ene generation to selectively identify protein interactions on cell membranes, a method called MicroMap (μMap). A light-triggered labeling technique that improves the spatial resolution of this type of mapping. Specifically, they relied on a photocatalyst with a very short range of energy transfer to activate a carbine-based tag that could only diffuse a small distance in water before the reaction.
Using photocatalytic-antibody conjugates to spatially localize carbene generation, they demonstrated selective labeling of antibody-bound targets and their microenvironment protein neighbors.
They used this technique to identify the constituent proteins of the programmed death-ligand 1 (PD-L1) microenvironment in living lymphocytes and selectively label them in immune synaptic junctions. Therefore, it is a valuable system in cancer immunotherapy.
Reference
David W. C. MacMillan et al. Microenvironment mapping via Dexter energy transfer on immune cells. Science. 2020. DOI: 10.1126/science.aay4106